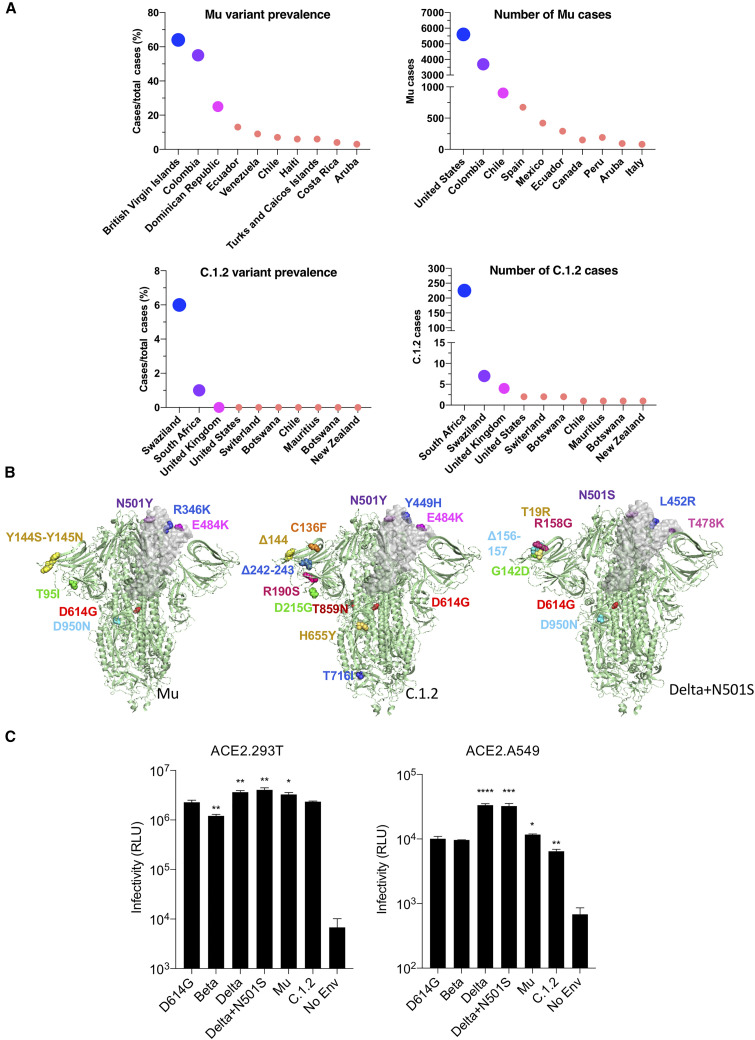
Figure 1.
Mu (B.1.621), C.1.2, and Delta+501S variant prevalence and spike protein mutations
(A) The global prevalence of Mu and C.1.2 variants is shown for countries with the highest prevalence or cases (extracted from https://outbreak.info/).
(B) Mutations in Mu, C.1.2, and Delta+N501S variant spikes are shown on the three-dimensional spike protein structure. A single RBD in each is shown in gray (side view). The Protein Data Bank (PDB) file of spike protein (PDB: 7BNM) (Benton et al., 2021) was downloaded from the PDB. 3D view of protein was obtained using PyMOL.
(C) The infectivity of Beta, Delta, Delta+N501S, Mu, and C.1.2 (-T716I) variant spikes pseudotyped lentiviruses on ACE2.293T and ACE2.A549 cells is shown. The viruses were normalized for RT activity and measured in triplicate with error bars that indicate the standard deviation. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. The experiment was done three times with similar results.