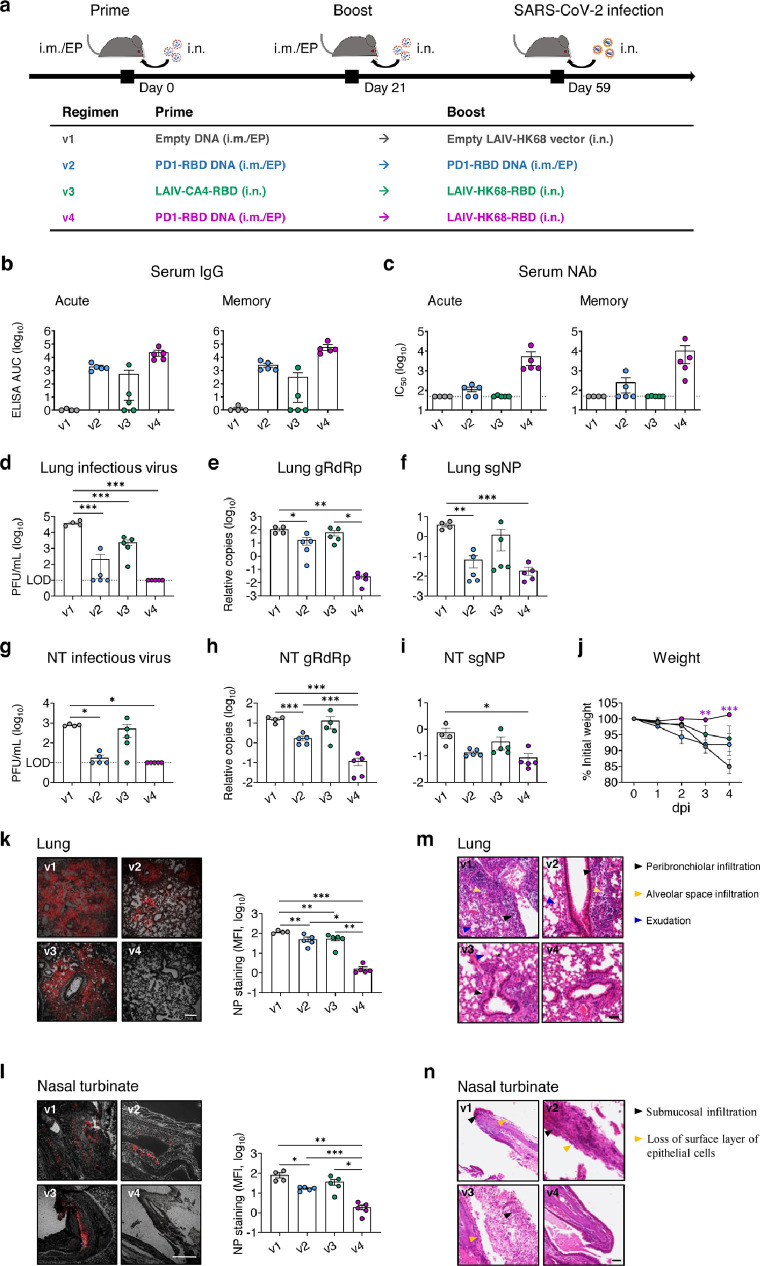
Figure 4.
PD1-RBD-DNA/LAIV-HK68-RBD regime prevents SARS-CoV-2 infection in the upper and lower respiratory tracts of hACE2 transgenic mice. (a) Experimental schedule and grouping of K18-hACE-2 mice (4 mice in v1 group and 5 mice in other groups). (b, c) Serum samples were collected for detection of anti-RBD IgG (b) and neutralizing antibody (c) against pseudovirus, respectively, on day 9 (acute, left) and day 33 (memory, right) post the 2nd immunization. The area under the curve (AUC) represents the total peak area calculated from ELISA OD values. A viral plaque assay was used to quantify infectious viruses in lung (d) and NT (g) homogenates. Log10-transformed plaque-forming units (PFU) per mL of tissue extractions were shown for each group. LOD: limit of detection. Sensitive RT PCR was used to quantify SARS-CoV-2 RdRp RNA (e, h) and NP subgenomic RNA (f, i) copy numbers (normalized by β-actin) in lung and NT homogenates. (j) Daily body weight was measured after infection. Differences between groups that were given the different vaccine regimes versus PBS were determined using a 2-tailed Student's t-test. Confocal images showed SARS-CoV-2 NP positive (red) cells in lungs (k) and NT (l) in the bright field. Scale bars: 200 µm. Mean fluorescent intensities (MFI) of NP+ cells in lung and NT were measured using ImageJ software and plotted with GraphPad prism. Representative images of animal lung (m) and NT (n) tissues by H&E. Scale bars: 50 µm. Each symbol represents an individual mouse with consistent color-coding. Error bars indicate the standard error of the mean. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. *p<0.05; **p<0.01; ***p<0.001.