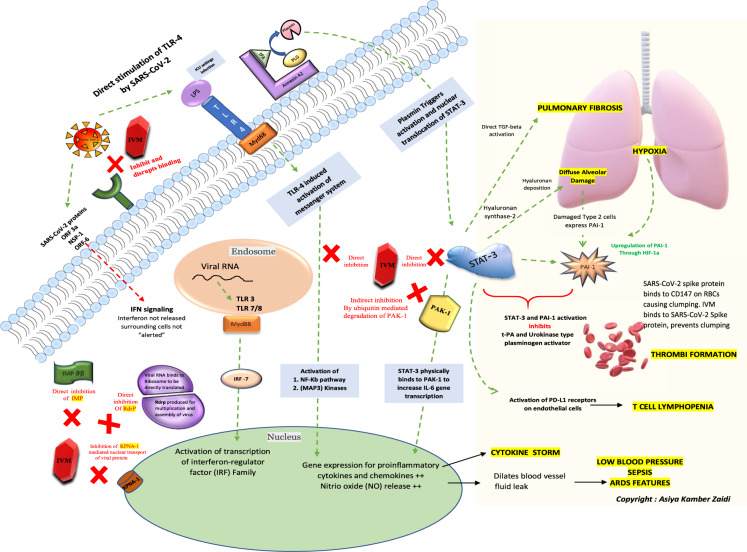
Fig. 1.
A schematic of the key cellular and biomolecular interactions between ivermectin, host cell, and SARS-CoV-2 in COVID-19 pathogenesis and prevention of complications: ivermectin (IVM) (red block) inhibits and disrupts binding of the SARS-CoV-2 S protein at the ACE-2 receptors (green). The green dotted lines depict activation pathways and the red dotted lines depict the inhibition pathways. The TLR4 receptors are directly activated by SARS-CoV-2 and also by LPS mediated activation (seen during ICU settings) causing activation of NF-Kb pathway and MAP3 kinases leading to increased intranuclear gene expression for proinflammatory cytokines and chemokines (responsible for cytokine storm) and NO release (responsible for blood vessel dilatation, fluid leak, low blood pressure, ARDS and sepsis). The NF-Kb and STAT-3 pathway activation is central to the pathogenesis and sequelae of COVID-19. STAT-3 physically binds to PAK1 and increases IL-6 transcription. The annexin A2 at the cell surface converts plasminogen; PLG to plasmin under the presence of t-PA. Plasmin triggers activation and nuclear translocation of STAT-3. An upregulation of STAT-3 stimulates hyaluronan synthase 2 in the lung cells causing hyaluronan deposition leading to diffuse alveolar damage and hypoxia. STAT-3 also directly activates TGF-beta initiating pulmonary fibrosis; a typical characteristic of SARS-CoV-2 lung pathology. The damaged type 2 cells express PAI-1 and an already hypoxic state also causes an upregulation of PAI (through hypoxic inducible factor-1) along with direct stimulation by STAT-3. Simultaneous STAT-3 and PAI-1 activation inhibits t-PA and urokinase-type plasminogen activator leading to thrombi formation. Also, the SARS-CoV-2 spike protein binds to the CD147 on red blood cells and causes clumping. IVM in turn binds to SARS-CoV-2 Spike protein and hence prevents clumping. T cell lymphopenia in COVID-19 can also be attributed to the direct activation of PD-L1 receptors on endothelial cells by STAT-3. IVM directly inhibits the NF-kb pathway, STAT-3, and indirectly inhibits PAK1 by increasing its ubiquitin-mediated degradation. The natural antiviral response of a cell is through interferon regulatory genes and viral RNA mediated activation of TLR-3 and TLR7/8- Myd88 activation of transcription of interferon regulator (IRF) family. For a virus to establish an infection, this antiviral response needs to be inhibited by blocking interferon production. The proteins such as importin and KPNA mediate nuclear transport of viral protein and subsequent IFN signaling. The SARS-CoV-2 proteins (ORF3a, NSP1, and ORF6) directly block IFN signaling causing the surrounding cells to become unsuspecting victims of the infection. IVM inhibits both importin ab (green) as well as the KPNA1 receptors (brown) causing natural antiviral IFN release. IVM also inhibits viral RdrP, responsible for viral replication. ACE-2 angiotensin-converting enzyme 2, LPS lipopolysaccharide, TLR Toll-like receptor, t-PA tissue-like plasminogen activator, PLG plasminogen, IMPab importin alpha-beta, Rdrp RNA dependant RNA polymerase, KPNA1 karyopherin subunit alpha 1, NF-kB nuclear factor kappa-light-chain-enhancer of activated B cells, Map3 kinases mitogen-activated kinases, PAK1 P21 activated kinase 1, STAT-3 signal transducer and activator of transcription 3, PAI-1 plasminogen activator inhibitor-1, HIF-1 hypoxia-inducible factor