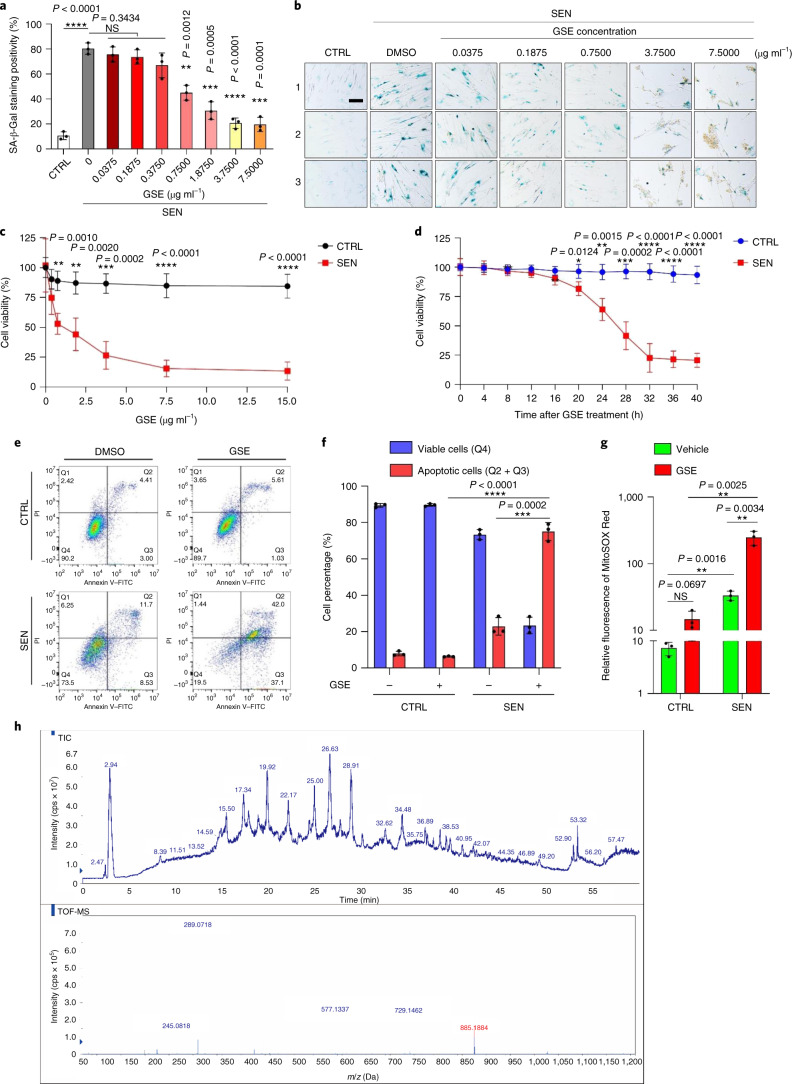
Fig. 1. Characterisation of the capacity of GSE to eliminate senescent cells.
a, Quantification of senescent PSC27 cell survival by SA-β-Gal positivity. GSE was applied to the medium at increasing concentrations. CTRL, control (proliferating) cells; SEN, senescent cells; NS, not significant. P values were calculated by one-way ANOVA with Tukey’s multiple-comparison test. b, Representative images displaying SA-β-Gal staining after treatment of PSC27 cells with different concentrations of GSE. Scale bar, 20 μm. Data are representative of three independent experiments. DMSO, dimethylsulphoxide. c, Survival analysis of control and senescent PSC27 cells upon treatment with GSE (at concentrations of 0.3750, 0.7500, 1.8750, 3.7500, 7.5000 and 15.0000 μg ml−1, respectively). Data are shown as mean ± s.d. and were derived from three biological replicates (n = 3 independent assays). P values were calculated by two-sided t-tests. d, Time course measurement of in vitro viability upon treatment of control and senescent PSC27 cells with GSE (3.75 μg ml−1). Data are shown as mean ± s.d. and were derived from three biological replicates (n = 3 independent experiments). P values were calculated by two-sided t-tests. e, Flow cytometry measurement of control and senescent PSC27 cells after processing with an annexin V–FITC and propidium iodide (PI) kit and 4,6-diamidino-2-phenylindole (DAPI) staining to determine the extent of apoptosis. Q1–Q4, quartiles 1–4. f, Comparative quantification of the percentage of viable (Q4, PI−annexin V−) and apoptotic (Q2 and Q3, PI+annexin V+ and PI−annexin V+, respectively) cells in control or senescent populations treated with vehicle or GSE for 3 d (n = 3 biologically independent assays). P values were calculated by two-sided t-tests. g, Measurement of the fluorescence signal of MitoSOX Red, a mitochondrial superoxide indicator, in PSC27 cells under different conditions. P values were calculated by two-sided t-tests. h, High-resolution mass spectra showing the total ion chromatogram (TIC) and base peak chromatogram of GSE after performing HPLC–ESI-QTOF-MS. Unless otherwise indicated, cells were subjected to relevant analyses 3 d after GSE treatment in the culture condition. cps, counts per second. Data in bar graphs and regression curves are shown as mean ± s.d. and are representative of three biological replicates. NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.