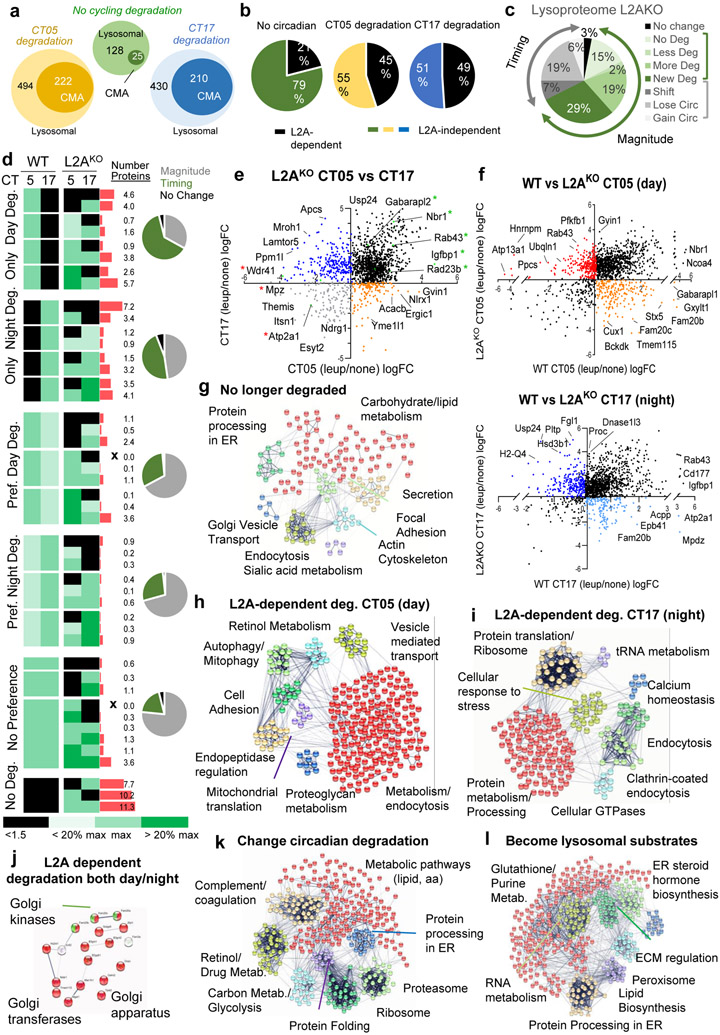
Fig. 8. CMA contributes to circadian remodeling of the proteome.
Comparative proteomics of liver lysosomes from leupeptin injected or not wild-type (WT) or LAMP2A knockout mice (L2AKO) isolated at circadian time CT05 (day) or CT17 (night). n=3 mice per CT, genotype and treatment group throughout the figure. a, Number of proteins undergoing lysosomal degradation (increase >1.5 folds after leupeptin treatment) or degradation through CMA (no longer increase upon leupeptin treatment in L2AKO mice) with preferential degradation or not at CT05 or CT17. b, Percentage of proteins degraded in a L2A-dependent (CMA) or -independent manner. c, Percentage of lysosomal proteins changing in magnitude or timing of degradation in L2AKO mice compared to WT. d, Heatmap of the differences in lysosomal degradation rates and timing between WT and L2AKO mice. Black: absence of degradation. Green color gradient: maximal degradation time and changes in >20% from maximal degradation. Red bars: percentage of total proteins in each group. X: no proteins in a group. Pie charts: percentage of proteins with differences in magnitude or time of degradation in each group in L2AKO mice. e, Log2 fold change (Log2FC) in rates of lysosomal degradation for individual proteins during the day and night cycle in L2AKO mice. Black: no cycle preference. Orange and blue: degraded only during the day or night, respectively. Grey: no longer degraded. Labels show examples of proteins in each group. f, Rates of lysosomal degradation (in LogFC) during the day (top) or night (bottom) in WT against L2AKO mice. Black: Proteins with preserved degradation time. Color: proteins displaying shifts in degradation time. g-l, STRING analysis for proteins no longer degraded in lysosomes in L2AKO mice (g), proteins displaying L2A-dependent degradation during the day (h) or during the night (i), proteins showing LAMP2A-dependent degradation both at CT05 and CT17 (j), proteins that change the time of their degradation upon LAMP2A ablation (k) and proteins degraded only in lysosomes from L2AKO mice but not from WT mice (l). All GO terms are statistically enriched with p<0.001. Numerical source data, statistics and exact p values are available as source data.