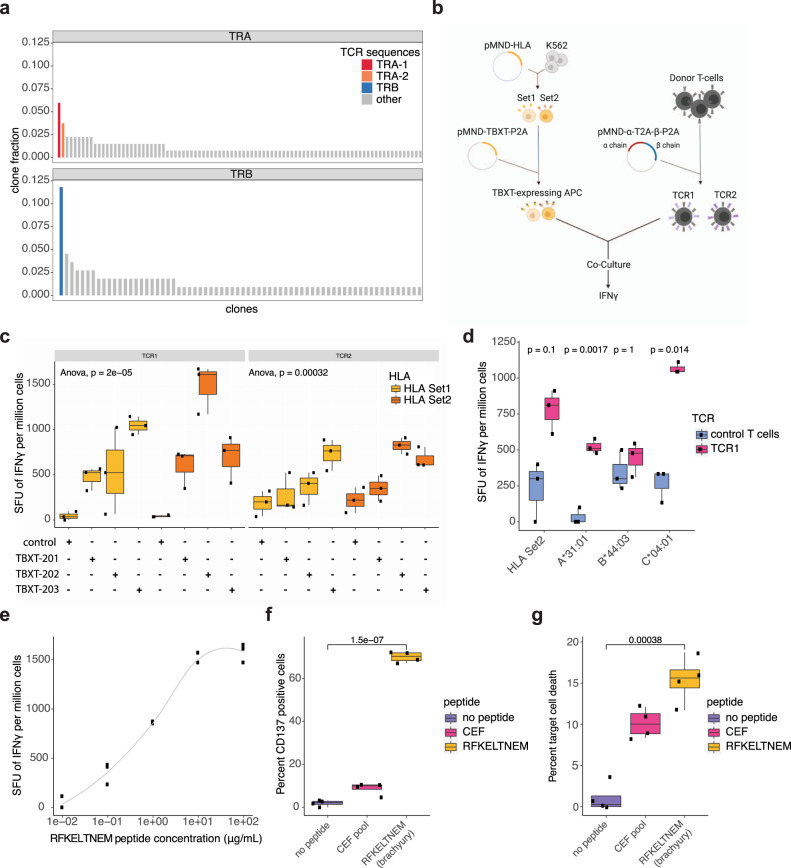
Fig. 4. Patient-derived TCR activation and cell killing in response to a brachyury peptide antigen.
a Unique TCR alpha (TRA) and TCR beta (TRB) chain sequences were inferred using bulk tumour RNA sequencing data from patient 2. The relative fraction of each sequence compared to all TRA and TRB aligned sequences is plotted on the y-axis. The two dominant TRA sequences TRA-1 and TRA-2 (red and orange bars, respectively) and the dominant TRB sequence (blue bar) were used for subsequent in vitro T cell activation ELISpot assays. b Schematic of ELISpot analysis of two reconstituted T cells (TCR1 and TCR2) expressing a patient-derived TRB chain and either of the two TRA chains, and synthetic antigen-presenting cells (APCs) transduced with one set of the patient’s classical HLA class I alleles (Set1 and Set2) and TBXT. IFNγ production is measured by the number of spot-forming units (SFU) per million cells. ELISpot results are shown for c TCR1 and TCR2 co-cultured with HLA Set1 or Set2 APCs expressing TBXT-201, 202, 203 or control (n = 3 for each condition, one way ANOVA; p values: TCR1 p = 2e−05 and TCR2 p = 0.00032). d TCR1 or control T cells and either HLA Set2 or HLA-A*31:01, HLA-B*44:03 or HLA-C*04:01 alleles (n = 3 for all conditions, unequal variance, two-sided T test Bonferroni corrected; p values for HLA Set2 p = 0.1, HLA-A*31:01 p = 0.0017, HLA-B*44:03 p = 1 and HLA-C*04:01 p = 0.014); e varying concentration of brachyury peptide RFKELTNEM with HLA-C*04:01 APCs with TCR1 T cells. Percentage of f CD137-positive T cells and g target cell death following co-culture with no peptide, HLA class I control CEF peptide pool or the RFKELTNEM APCs and TCR1 T cells (n = 4 for all conditions, unequal variance, two-sided T test shows a significant difference between no peptide and RFKELTNEM in f, g). Box plots in c, d, f, g represent median, upper and lower quartiles, and whiskers represent limits of the distributions (1.5-times interquartile range).