Figure 4.
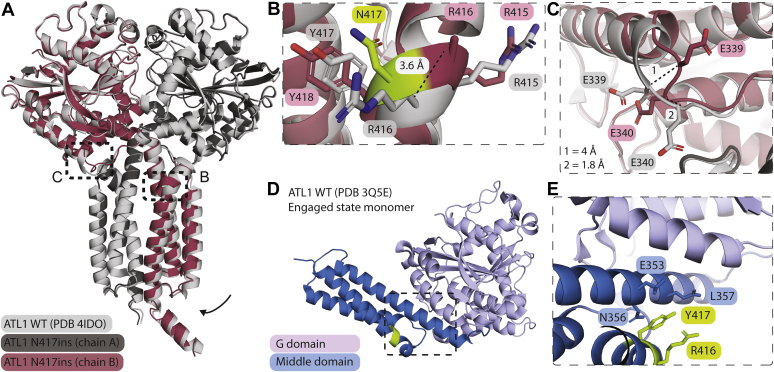
Structure of the ATL1 N417ins crossover dimer bound to GDP•AlF4−.A, structural overview. ATL1 N417ins (dark gray and maroon) superimposed with ATL1 WT (light gray; PDB 4IDO) each bound to GDP•AlF4− and Mg2+. Black boxes indicate boundaries of zoom-in views in B and C. Arrow indicates slight rotation of mutant middle domain. B, zoom-in view of the insertion mutation in chain B. Each residue is labeled (number of N417ins residues were adjusted after residue N417 to accommodate the extra residue). The distance spanning the corresponding R416 Cα positions between the WT and N417ins structures is indicated (3.6 Å). N417 is colored in green while other residues were colored as in A. C, magnified view of the transition between the G domain and linker region of chain B, with both E339 and E340 of each structure labeled. Distances between the same residue in both structures is indicated in the lower left corner. D, structure of the monomeric, engaged ATL1 WT state (PDB 3Q5E) bound to GDP and Mg2+. The residues directly pre- and proceeding the N417ins mutation (R416 and Y417) are indicated in green. The black box indicates the region shown in E. E, position of the insertion mutation. R416 and Y417 lie directly adjacent to middle and G domain helices integral in engaged state formation.