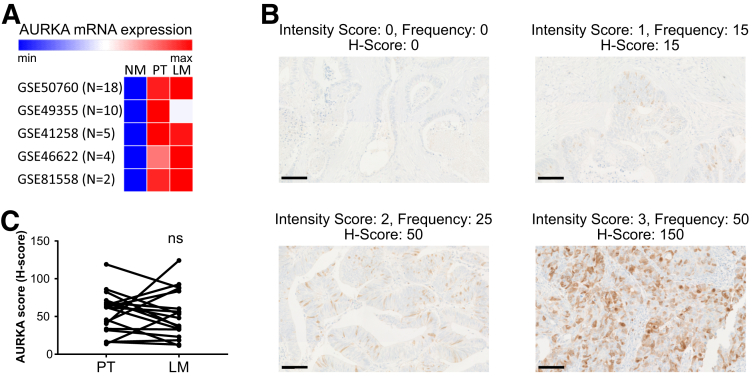
Figure 10.
Expression of AURKA in patient-matched cases of primary tumors and their liver metastasized derivatives. (A) Heat map showing AURKA mRNA expression in 5 different CRC cohorts containing patient-matched tissue samples of normal mucosa (NM), primary tumor (PT), and liver metastasis (LM). Gene Expression Omnibus database accession numbers of the analyzed CRC cohorts are indicated. (B) Immunohistochemistry for detection of AURKA in FFPE CRC tissue sections. Staining within tumor areas was quantified with respect to intensity (0–3 grading scale: 0 = no staining, 1 = weak staining, 2 = moderate staining, 3 = strong staining), and frequency (0%–100% of AURKA-positive cells in 5% increments). The H-score (possible range, 0–300) was obtained by multiplying staining intensity and the proportion of positive cells in a given area. Scale bars: 100 μm. (C) AURKA protein abundance (H-scores) as detected by immunohistologic staining on a CRC cohort (n = 18 pairs) of primary tumors and their patient-matched liver metastases. Statistical significance was assessed by a paired t test (ns, non-significant: P = .830). max, maximum; min, minimum.