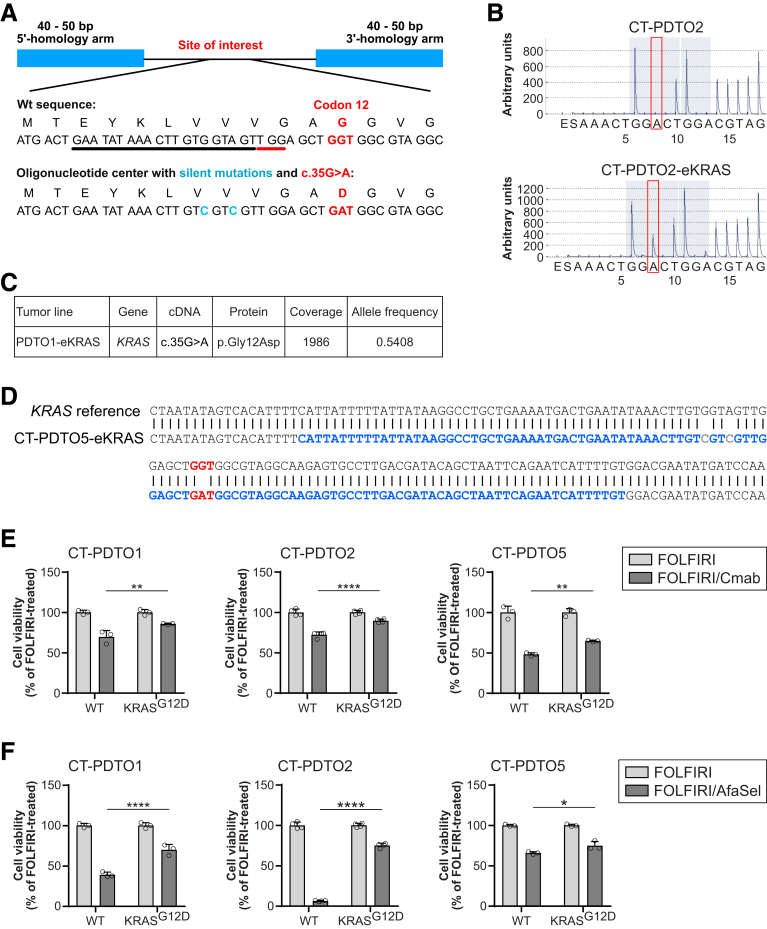
Figure 7.
CRISPR/Cas9-mediated engineering of KRASG12Din FOLFIRI/Cmab-tolerant CRC organoids. (A) Schematic representation of the repair oligonucleotide provided to PDTO cells together with CRISPR/Cas9-ribonucleoparticles targeting KRAS exon 2. Black underline indicates the 20-mer single guide RNA target sequence, the protospacer adjacent motif is shown with red underline. The oncogenic GGT>GAT (c.35G>A) mutation is shown in red. Silent mutations in the repair oligonucleotide are indicated in blue. (B) Analysis of mutations in the KRAS gene by using pyrosequencing technology. Red frames indicate the DNA nucleotide, which is mutated in oncogenic KRAS. Note the appearance of this A (GGT>GAT) only case of the CRISPR/Cas9-edited PDTO2 (PDTO2–eKRAS, lower panel). (C) Exemplary result of panel sequencing on PDTO1–eKRASG12D. The KRAS locus of interest was sequenced 1986 times (coverage), and 54.08% of sequences showed the G12D-encoding variant (allele frequency). (D) Sanger sequencing of the edited KRAS locus. Repair oligo sequence is indicated in blue. Oncogenic KRAS (GGT>GAT) mutation is highlighted in red, introduced silent mutations are indicated in grey. (E and F) Analysis of cell viability (CellTiter Glo 3D) on the indicated PDTO cultures treated with FOLFIRI alone or in combination with either (A) Cmab or (B) afatinib (dual EGFR/HER2 inhibitor) plus selumetinib (MEK inhibitor) (AfaSel). Statistical significance of the differential treatment effects between PDTO lines harboring either wild-type or oncogenic KRAS was assessed by 2-way ANOVA plus the Sidak multiple comparisons test and is indicated by asterisks (∗P ≤ .05, ∗∗P ≤ .01, ∗∗∗∗P ≤ .0001). Means ± SD, n = 3. Wt, wild-type.