Fig. 4.
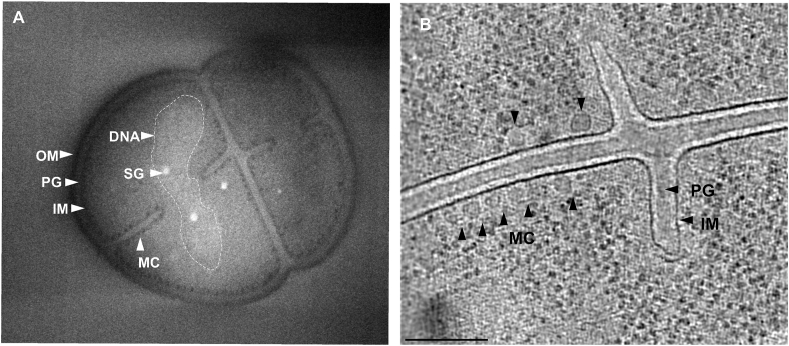
3D volume imaging vs cryo-ET for detecting subcellular structures. A) Cryo-FIB-SEM volume imaging revealed the overall cell shape of dividing cells. The cell envelope architecture was poorly discerned. Numerous macromolecular complexes (MC) associated with the IM were clearly visible. Storage granules (SG) were associated with segregating DNA (dashed line). Scale bar, 500 nm. B) A slice through the center of a cryotomogram generated using SIRT-like reconstruction and Topaz-Denoise algorithm clearly revealed the PG (∼40 nm) and an invaginating IM during cell division. Some MC along the IM (black arrows) were also apparent. Scale bar, 200 nm.