Abstract
Cellular senescence is a complex stress response defined as an essentially irreversible cell cycle arrest mediated by the inhibition of cell cycle-specific cyclin dependent kinases. The imbalance in redox homeostasis and oxidative stress have been repeatedly observed as one of the hallmarks of the senescent phenotype. However, a large-scale study investigating protein oxidation and redox signaling in senescent cells in vitro has been lacking. Here we applied a proteome-wide analysis using SILAC-iodoTMT workflow to quantitatively estimate the level of protein sulfhydryl oxidation and proteome level changes in ionizing radiation-induced senescence (IRIS) in hTERT-RPE-1 cells. We observed that senescent cells mobilized the antioxidant system to buffer the increased oxidation stress. Among the antioxidant proteins with increased relative abundance in IRIS, a unique 1-Cys peroxiredoxin family member, peroxiredoxin 6 (PRDX6), was identified as an important contributor to protection against oxidative stress. PRDX6 silencing increased ROS production in senescent cells, decreased their resistance to oxidative stress-induced cell death, and impaired their viability. Subsequent SILAC-iodoTMT and secretome analysis after PRDX6 silencing showed the downregulation of PRDX6 in IRIS affected protein secretory pathways, decreased expression of extracellular matrix proteins, and led to unexpected attenuation of senescence-associated secretory phenotype (SASP). The latter was exemplified by decreased secretion of pro-inflammatory cytokine IL-6 which was also confirmed after treatment with an inhibitor of PRDX6 iPLA2 activity, MJ33. In conclusion, by combining different methodological approaches we discovered a novel role of PRDX6 in senescent cell viability and SASP development. Our results suggest PRDX6 could have a potential as a drug target for senolytic or senomodulatory therapy.
Keywords: Cellular senescence, Interleukin 6, Peroxiredoxin 6, Redox proteomics, Senescence-associated secretory phenotype, SILAC-iodoTMT
Graphical abstract
Highlights
-
•
SILAC-iodoTMT is a powerful tool to quantify redox imbalance in IRIS.
-
•
Senescence in hTERT-RPE-1 cells is not accompanied by bulk cysteine oxidation.
-
•
Antioxidant proteins are upregulated in senescent hTERT-RPE-1 cells.
-
•
PRDX6 silencing affects redox homeostasis and viability of senescent cells.
-
•
PRDX6 silencing alters secretome of senescent RPE-1 cells and suppresses IL-6.
Abbreviations
- AA
arachidonic acid
- ACN
acetonitrile
- BrdU
5-bromo-2′-deoxyuridine
- Ctrl
control
- Cp
peroxidatic cysteine
- Cys
cysteine
- DAMPs
damage-associated molecular patterns
- DAPI
4′,6-diamidino-2-phenylindole
- dFBS
dialyzed fetal bovine serum
- DMEM
Dulbecco's Modified Eagle Medium
- EDTA
ethylenediaminetetraacetic acid
- EdU
5-ethynyl-2′-deoxyuridine
- FA
formic acid
- FBS
fetal bovine serum
- f.c.
final concentration
- FDR
false discovery rate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GRX1
Glutaredoxin 1
- H2O2
hydrogen peroxide
- HBSS
Hank's balanced salt solution
- iodoTMT
iodoacetyl tandem mass tag
- iPLA2
phospholipase A2
- IR
ionizing radiation
- IRIS
ionizing radiation-induced senescence
- IW
isolation window
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LPCAT
lysophosphatidylcholine acyl transferase
- NCE
normalized collision energy
- NH4FA
ammonium formate
- OIS
oncogene-induced senescence
- PBS
phosphate buffered saline
- PCA
principal component analysis
- Prx
peroxiredoxin
- ROS
reactive oxygen species
- RT
room temperature
- SASP
senescence-associated secretory phenotype
- SDC
sodium deoxycholate
- SILAC
stable isotope labeling with amino acids in cell culture
- SIPS
stress-induced premature senescence
- TCEP
tris(2-carboxyethyl) phosphine hydrochloride
- TEAB
triethylammonium bicarbonate
- TFA
trifluoroacetic acid
- TMRE
tetramethylrhodamine ethyl ester perchlorate
1. Introduction
Cellular senescence is a complex cellular stress response triggered by molecular damage and characterized by a prolonged and generally irreversible cell cycle arrest. This stable arrest, and more importantly, the ability of senescent cells to influence their microenvironment, contribute to their role in the development of age-related pathologies including cancer [1,2]. Various types of stresses including DNA damage and oxidative stress are responsible for expression of protein inhibitors of cyclin-dependent kinases, the pivotal effectors of cellular senescence. The evidence of mechanistic role of oxidative stress in the development of cellular senescence has been laid by the work of Toussaint et al. [3]. The authors showed that in vitro exposure of cells to oxidants such as hydrogen peroxide (H2O2) resulted in proliferation arrest associated with replicative senescence-like morphological changes, termed stress-induced premature senescence (SIPS) [3]. There has also been accumulating evidence that oxidative stress is involved in the onset of replicative senescence [4,5], oncogene-induced senescence (OIS) [6,7], and ionizing radiation-induced senescence (IRIS) [8]. Furthermore, application of antioxidants can mitigate senescence development [8,9]. It has been proposed that deregulation of NADPH oxidases [[10], [11], [12], [13]] or mitochondrial dysfunction [[14], [15], [16]] are the internal sources of reactive oxygen species (ROS) responsible for the development of cellular senescence. Thus, redox homeostasis imbalance and prolonged oxidative stress have been repeatedly observed as one of the major components of the senescent phenotype.
Oxidative stress can lead to damage of all macromolecules including nucleic acids, proteins, lipids and their precursors, and metabolites. Thus, all cells must be equipped with cytoprotective antioxidant enzymes. Among them, the peroxiredoxin (Prx) family of peroxidases [17,18] has been shown to maintain cellular redox homeostasis and viability [19], acting as H2O2-scavenging enzymes, but also to contribute to redox signaling as redox relays [20] or functioning as floodgates [21]. PRDX6 is a unique 1-Cys peroxiredoxin family member [18,22] crucial for maintenance of phospholipid homeostasis due to its unique ability to bind and reduce phospholipid hydroperoxides. Additionally, it has been described to exert phospholipase A2 (iPLA2) [23,24] and lysophosphatidylcholine acyl transferase (LPCAT) activities [25]. The regulation of the PRDX6 enzymatic functions occurs at multiple levels – by subcellular localization, protein-protein interactions, and post-translational modifications [26]. Perhaps most importantly, the irreversible hyperoxidation of the peroxidatic cysteine (Cp)47 has been described as a regulator of the iPLA2 activity of PRDX6 [27]. A number of studies have demonstrated that PRDX6 expression provides protection from ROS of multiple origins in cancer and normal cells, both in vivo and in vitro, prevents lipid peroxidation damage [28], cell death [[29], [30], [31]], and ensures normal proliferation and redox regulation of metabolic pathways [32,33]. Similarly to other members of the Prx family, PRDX6 has been shown to play multifaceted roles in inflammatory processes [34]. While the antioxidant function of PRDX6 can be mostly considered as anti-inflammatory, its iPLA2 activity and the ability to release arachidonic acid (AA) has been linked to inflammation in experiments utilizing the iPLA2 activity inhibitor, MJ33 [28,35,36]. In addition to the iPLA2 activity, peroxiredoxins, including PRDX6, can be released to extracellular space where they might function as damage-associated molecular patterns (DAMPs [37]) activating the pro-inflammatory pathways via the TLR4 receptor [38]. There is a vast research mapping the physiological functions of PRDX6 in multiple organs or cell types such as lungs, brain, eye, or spermatozoa and also pathophysiological functions implying the role of PRDX6 in tumor viability [[29], [30], [31]], progression, and metastasis [[39], [40], [41], [42], [43]]. It has been shown that PRDX6 protects normal cells from oxidative damage [44,45] and cellular senescence [46,47]. However, to the best of our knowledge, the role of PRDX6 in the development of the senescent phenotype has not been addressed yet.
In this study, we employed our recently published SILAC-iodoTMT methodology to detect and quantitatively estimate the level of protein sulfhydryl oxidation and proteome-level changes [48] in the IRIS in vitro model of hTERT-RPE-1 cells suffering from elevated ROS formation. We identified PRDX6 as an important player in cellular redox homeostasis in IRIS. Subsequently, we applied MS-based proteomic strategy and siRNA-mediated knock-down of PRDX6 to evaluate the contribution of PRDX6 to the senescent phenotype. We described that the lack of PRDX6 activity during the development of IRIS led to suppression of important phenotypic features of senescence as well as senescence-associated secretory phenotype (SASP) leading to altered composition of extracellular matrix, protein secretion, and more specifically, decreased secretion of pro-inflammatory cytokine IL-6.
2. Material and methods
2.1. Reagents and chemicals
All reagents including antibodies, siRNAs, and primer sequences used in this study are provided in Supplementary Material and Methods section.
2.2. Cell culture
Human telomerase-immortalized retinal pigment epithelial cells hTERT-RPE-1 (ATCC CRL-4000) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco's Modified Eagle Medium (DMEM) with 4.5 g/L glucose and 4 mM l-glutamine supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL). Human embryonal lung fibroblasts MRC-5 cells (ATCC CLL-171) were cultured in DMEM with 1 g/L glucose and 4 mM l-glutamine supplemented with 10% FBS, penicillin (100 U/mL), streptomycin (100 μg/mL), and non-essential amino acids.
2.3. Cell culture SILAC-iodoTMT
hTERT-RPE-1 were cultured as described previously [48]. DMEM was supplemented with 200 mg/L l-proline, 10% dialyzed fetal bovine serum (dFBS), 0.1% FBS, penicillin (100 U/mL), and streptomycin (100 μg/mL). SILAC labeling was done by using l-arginine [13C6, 15N4] and l-lysine [13C6] for more than six cell population doublings, and the incorporation efficiency was monitored throughout the cultivation. Cell cultures were maintained at 37 °C under 5% CO2 atmosphere and 95% humidity.
2.4. SDS-PAGE and immunoblotting
SDS-PAGE and immunoblotting were performed according to standard protocols. Samples (20–30 μg) in a reducing sample buffer were separated by SDS-PAGE on a 12% gel and detected by immunoblotting. GRX1 detection was performed using Tricine SDS-PAGE [49]. Nitrocellulose membranes were blocked in 5% milk at room temperature (RT) for 1 h, incubated with a primary antibody (1 : 1000) at 4 °C overnight and after washing three times with phosphate buffered saline (PBS)/Tween-20, incubated with a secondary antibody (1 : 10,000) at RT for 1 h (anti-mouse/anti-rabbit IgG HRP, 1 : 10,000; BioRad, Hercules, CA, USA). Proteins were visualized using chemiluminescence (Amersham TM, GE Healthcare) and analyzed for relative band densities (ImageJ software). The list of antibodies used in this study is provided in the Supplementary Material and Methods section.
2.5. Detection of senescence-associated activity of beta-galactosidase
The detection of senescence-associated activity of beta-galactosidase was performed as described previously [50]. Briefly, hTERT-RPE-1 cells were seeded at low density (5000 cells/cm2) and irradiated after 16 h by a dose of 20 Gy. Eight days after irradiation and two days after seeding of proliferating control cells, the cells were fixed using 0.5% glutaraldehyde (15 min, RT) and incubated with the X-gal staining solution at 37 °C until blue color appeared in the senescent cell culture.
2.6. 5-Ethynyl-2′-deoxyuridine incorporation assay
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay was performed using Click-iT™ EdU Cell Proliferation Kit (#C10337, ThermoFisher Scientific). hTERT-RPE-1 cells were seeded at low density (5000 cells/cm2) and after 16 h irradiated by a dose of 20 Gy. Eight days after irradiation and two days after seeding of a proliferating control cells (seeded 10,000 cells/cm2), 10 μM EdU was added, and the cells were cultured for 24 h. The cells were fixed using 4% formaldehyde (15 min, RT) and permeabilized using 0.5% Triton X-100 in PBS (20 min, RT). The click reaction was performed according to the manufacturer's protocol, and the nuclei were counterstained using 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) solution (2 min, RT).
2.7. Fluorescent microscopy – DNA damage foci
hTERT-RPE-1 cells were seeded at low density (5000 cells/cm2) and after 16 h irradiated by a dose of 20 Gy. Eight days after irradiation and two days after seeding of a proliferating control cells (seeded 10,000 cells/cm2), the cells were fixed using 4% formaldehyde (10 min, RT) and permeabilized using 0.2% Triton X-100 in PBS (10 min, RT). After blocking of non-specific binding using DMEM/10% FBS (30 min, RT), the coverslips were incubated with a primary antibody (1 : 200, 1 h, RT) diluted in blocking solution, following by incubation with the secondary antibody (1 : 1,000, 16 h, 4 °C) in blocking solution. Nuclei were counterstained using 1 μg/mL DAPI solution (2 min, RT).
2.8. Real-time quantitative reverse transcription PCR
hTERT-RPE-1 cells were seeded at a density of 20,000 cells/cm2 and after 16 h irradiated by a dose of 20 Gy [51]. Eight days after irradiation and two days after seeding of proliferating control cells (seeded 10,000 cells/cm2), cells were harvested, and RNA was isolated using RNeasy Mini Kit (#74106, Qiagen) according to the manufacturer's protocol. Contaminating DNA was removed by RNase-Free DNase Set (#79254, Qiagen) on RNA easy column according to the manufacturer's protocol. cDNA was synthesized using a MultiScribe™ Reverse Transcriptase kit (#4311235, ThermoFisher Scientific). Quantitative reverse transcription PCR was performed in ABI Prism 7300 (Applied Biosystems) using SYBR™ Select Master Mix (#4472919, ThermoFisher Scientific) with the primers shown in the Supplementary Material and Methods section. The relative quantity of cDNA was estimated by ΔΔCt [52], data were normalized to β-actin, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), or their average.
2.9. siRNA-mediated gene knock-down
Cells were transfected with 10 nM siRNA using Lipofectamine® RNAiMAX Transfection Reagent (#13778150; ThermoFisher Scientific) following the manufacturer's protocol. The siRNA sequences are shown in Supplementary Material and Methods section. Silencer™ Select Negative Control No. 1 siRNA (#4390843, ThermoFisher Scientific) was used as a negative control (siNC). For siRNA validation, the cells were seeded at a density of 15,000/cm2, transfected 16 h after seeding, and harvested after 48 h or 72 h after transfection for mRNA and protein analysis, respectively. For all senescent cells analyses, the cells were seeded at a density of 20,000/cm2 and irradiated (20 Gy) 16 h after seeding. The cells were first transfected 2 days after irradiation, the second transfection was performed 5 or 6 days after irradiation (both designs were validated to provide comparable results), and the cells were harvested for experiments 8 or 9 days after irradiation as indicated.
2.10. Detection of intracellular ROS, mitochondrial ROS and membrane potential, and lipid peroxidation
Intracellular ROS production was detected using CellROX™ Deep Red Reagent (#C10422; Thermo Fisher Scientific) according to the optimized manufacturer's protocol (30 min incubation with 5 μM dye dissolved in the cell culture medium at 37 °C). Mitochondrial ROS production was detected using MitoSOX™ Red Mitochondrial Superoxide Indicator (#M36008; Thermo Fisher Scientific) according to the optimized manufacturer's protocol (60 min incubation with 5 μM dye dissolved in cell culture medium at 37 °C). Lipid peroxidation was detected using BODIPY™ 581/591C11 (Lipid Peroxidation Sensor; #D3861; Thermo Fisher Scientific) according to optimized manufacturer's protocol (30 min incubation with 2.5 μM dye dissolved in the cell culture medium at 37 °C). Mitochondrial membrane potential was analyzed using Tetramethylrhodamine, Ethyl Ester, Perchlorate assay (TMRE; #T669; Thermo Fisher Scientific) according to the optimized manufacturer's protocol (15 min incubation with 0.5 μM dye dissolved in cell culture medium at 37 °C). In all assays, DMSO-treated cells were used as a control to subtract autofluorescence in the post-analysis. Cells were washed with PBS (37 °C), detached with 0.25% trypsin/EDTA, resuspended in PBS, 1 g/L glucose, 1% FBS and centrifuged (300×g, 3 min, 4 °C). Pellets were resuspended in 100 μL PBS, 1 g/L glucose, 1% FBS and analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA). Hoechst 33258 or propidium iodide were used to exclude non-viable cells from the analysis.
2.11. Detection of extracellular ROS production
Extracellular ROS were detected using Amplex™ Red Hydrogen Peroxide/Peroxidase Assay Kit (#A22188; Thermo Fisher Scientific) according to the manufacturer's protocol and a previously published procedure [53]. Briefly, the cells were detached by 0.25% trypsin/EDTA, counted, and 150,000 proliferating or senescent cells (20 Gy, 8 days after irradiation) were used for analysis. Resorufin production was monitored every minute for 60–90 min to estimate kinetics of H2O2 release from the cells. Light microscopy was used to estimate the diameter of the detached cells, and their volume was calculated under the assumption the detached cells have approximately a spherical shape.
2.12. Detection of apoptosis
Apoptosis assay was performed using annexin V/Hoechst 33258 staining. Briefly, cells in the cell culture supernatant were collected, and the adherent cells were detached using 0.25% trypsin/EDTA. Both fractions were pooled, and the cells were washed three times in 0.5 mL of 1 × Annexin binding buffer. Annexin V staining was performed in 100 μL of 1 × Annexin binding buffer with Annexin V Dyomics 647 antibody (1 : 100; Exbio, Vestec, Czech Republic) for 15 min on ice with no light exposure. Hoechst 33258 was added prior to the flow cytometry analysis (BD Biosciences, San Jose, CA, USA).
2.13. Viability assays
Cells were seeded in 96-well plates at density of 20,000 cells/cm2 (6440 cells per well). Proliferating or senescent cells (control, siNC, or siPRDX6 transfected as described above) were treated with H2O2 in triplicate in a concentration range 0.025–3.5 mM for proliferating cells and 0.5–10 mM for senescent cells for 24 h. To determine cell viability by the crystal violet assay [54,55], the cells were washed twice with 150 μL PBS and then stained in 30 μL 0.5% (w/v) crystal violet in 20% methanol for 15 min. Plates were washed three times with double distilled H2O and left to dry overnight. Crystal violet was solubilized with 75 μL 0.2% Triton X-100 (Sigma-Aldrich, St. Louis, MO) in PBS for 15 min. Absorbance of crystal violet was measured at 595 nm using a microplate reader (Multiskan EX, Thermo Electron Corporation, Waltham, MA). Alternatively, the MTT assay was performed with CellTiter 96® Non-Radioactive Cell Proliferation Assay kit (#G4000, Promega) according to the manufacturer's protocol. Absorbance of the treated samples was expressed as a percentage of absorbance of untreated cells. IC50 values were estimated using nonlinear regression curve fitting in GraphPad Prism version 8.0.0.
2.14. Inhibition of PRDX6 iPLA2 activity by MJ33
hTERT-RPE-1 cells were seeded at medium density (15,000 cells/cm2) and irradiated by a dose of 20 Gy 16 h after seeding. Six days after irradiation, 1-hexadecyl-3-trifluoroethylglycero-sn-2-phosphomethanol (MJ33; #M3315, Sigma-Aldrich), an iPLA2 activity inhibitor [56] was added in a final concentration of 10 μM, 25 μM, and 50 μM. The cells were incubated with the inhibitor for 72 h, and the inhibitor solution in cell culture medium was replaced every 24 h to ensure continuous treatment. Cell culture supernatant was collected on day 9 after irradiation and processed as described below.
2.15. Detection of cytokines in cell culture media
The cell culture supernatant was collected 24 h after changing the culture media for fresh. The medium was subjected to a two-step centrifugation to remove cells and debris contamination (200×g, 5 min, 4 °C; 2000×g, 5 min, 4 °C). Adherent cells were detached using 0.25% trypsin/EDTA, and the cell number was estimated using flow cytometry. The relative concentration of 11 cytokines was estimated using a Human Inflammation 11-Plex Panel kit (#C192211, AimPlex Biosciences, Inc., Pomona, CA, USA) according to manufacturer's protocol. Resulting relative fluorescence intensities were corrected for cell count and expressed as fold change relative to control or siNC.
2.16. Redox-proteomic analysis of senescent cells
The experiment was performed in four biological replicates with swapped SILAC groups, i.e., the experiment contained two “light” and two “heavy” samples from each condition (proliferating control cells and senescent cells). hTERT-RPE-1 cells were seeded at a density of 10,000 cells/cm2 and after 16 h irradiated by a dose of 20 Gy. Eight days after irradiation and two days after seeding of proliferating control cells (seeded 15,000 cells/cm2), the dishes were washed three times with ice-cold PBS and lysed with ice-cold lysis buffer (3% SDC/200 mM TEAB/1 mM EDTA). After the addition of the lysis buffer, the cells were scraped, transferred into microtubes, and the lysate was frozen in liquid nitrogen. The lysates were defrosted in an ice-cold water bath (4 °C) in BioRuptor and sonicated (low intensity, 30 s ON, 30 s OFF, three cycles). Remaining debris was removed by centrifugation (16,100×g, 10 min, 4 °C). During the whole procedure, the tubes were kept on ice and protected from light exposure, and whenever possible, the sample handling was performed in a cold room (4 °C).
2.17. Redox-proteomic analysis of senescent cells after PRDX6 silencing
The experiment was performed in two biological replicates per siRNA with swapped SILAC groups, i.e., the experiment contained two “light” and two “heavy” siNC samples, and one “light” and “heavy” samples from each siPRDX6. hTERT-RPE-1 cells were seeded at a density of 20,000 cells/cm2 and after 16 h irradiated by a dose of 20 Gy. Two sequential transfections were performed on day 2 and day 5 after irradiation as described above, and the cells were harvested 8 days after irradiation using the protocol described in the previous paragraph.
2.18. Sequential iodoTMT labeling, protein digestion and peptide desalting
Sequential iodoTMT labeling, protein digestion and peptide desalting were conducted as previously described [48]. Briefly, concentration of all cell lysate samples was adjusted to 1 μg/μL using the lysis buffer and 10 mM neocuproine (f.c. = 0.1 mM). The sample lysate (Condition 1; proliferating cells) labeled in the light/heavy SILAC channel (50 μg) was mixed with an equal amount of the sample lysate (Condition 2; senescent cells) of the complementary heavy/light SILAC group (Supplementary Fig. 1A). Free –SH groups were labeled by the first iodoTMT label (TMT1) at 37 °C for 2 h. Subsequently, proteins were precipitated using cold acetone (−20 °C) for 60 min. The pellet was re-dissolved in lysis buffer with 0.1 mM neocuproine and incubated with TCEP (f.c. = 5 mM) at 50 °C for 60 min to reduce the reversibly oxidized cysteines. The newly reduced cysteines were then labeled using the second iodoTMT label (TMT2) and the reaction was quenched by l-cysteine hydrochloride (f.c. = 20 mM). After the second acetone precipitation, proteins were digested using rLys-C (FUJIFILM Wako, Osaka, Japan) at 37 °C for 3 h followed by sequencing grade trypsin (Promega) digestion at 37 °C overnight at 1 : 50 ratio (enzyme-to-protein). Digestion was stopped by addition of trifluoroacetic acid (TFA, f.c. = 2%), and precipitated SDC was removed by extraction in water-saturated ethyl acetate. Finally, samples were desalted and evaporated to dryness. IodoTMTsixplex™ Isobaric Label Reagent Set for one simplex experiment (Thermo Scientific, Waltham, MA, USA) was used for peptide iodoTMT labeling.
2.19. Basic fractionation liquid chromatography
In the first dimension, 85 μg of each sample was re-dissolved in a mobile phase A (2% ACN, 20 mM NH4FA, pH 10) and 80 μg was injected for basic fractionation on UltiMate 3000 RSLC system (Thermo Fisher Scientific, Bremen, Germany) equipped with UV detection. Peptides were separated in a XBridge BEH column (2.1 μm × 150 mm, C18, 2.5 μm, 130 Å, from Waters, Milford, MA, USA) using a linear gradient of 3%–50% mobile phase B (80% ACN, 20 mM NH4FA, pH 10) at a flow rate 0.4 mL/min, at 40 °C, for 30 min. Peptide fractions were collected in the range of 0.5–37 min per 45 s. In total, 49 fractions were collected and 3rd – 42nd fractions were matched in the final set of 8 fractions (Supplementary Fig. 1C). Those were evaporated to dryness and stored at −80 °C for further analysis. All buffers and mobile phases for LC separation were prepared in LC-MS grade water purchased from Honeywell (Morris Plains, NJ, USA).
2.20. Nano-liquid chromatography coupled to tandem mass spectrometry analysis
Evaporated fractions were re-dissolved in a mobile phase A (0.1% TFA in 2% ACN) and 2 μg were injected onto UltiMate 3000 RSLCnano system (Thermo Fisher Scientific, Bremen, Germany) for the liquid chromatography separation. The analytical system consisted of PepMap 100C18, 3 μm, 100 Å, 75 μm × 20 mm trap column and PepMap RSLC C18, 2 μm, 100 Å, 75 μm × 500 mm analytical column (both from Thermo Fisher Scientific). The samples were loaded onto the trap column in 0.1% TFA in 2% ACN at 8 μL/min for 3 min. Tryptic peptides were separated by a segment gradient running from 2% to 9% of a mobile phase B (80% ACN with 0.1% FA) for 57 min, further from 9% to 34.5% of B for 160 min, and finally to 45% of B for 23 min at a flow rate of 200 nL/min. Eluted peptides were electrosprayed into Q-Exactive Plus using a Nanospray Flex ion source (both from Thermo Fisher Scientific, Bremen, Germany). Positive ion full scan MS spectra were acquired in the range of 350–1600 m/z using 3 × 106 AGC target in the Orbitrap at 70,000 resolution with a maximum ion injection time of 50 ms. Parameters of the isolation window (IW) and normalized collision energy (NCE) were set 1.6 m/z for IW and 30 for NCE as previously described [48]. MS/MS spectra were acquired at resolution of 35,000, with a 1 × 106 AGC target and a maximum injection time of 120 ms. Only 15 of the most intensive precursors with minimal AGC target of 2.4 × 104 and a charge state ≥2 were fragmented. The dynamic exclusion window was 17 s. The fixed first mass was set to 100 m/z and the scan range from 200 to 2000 m/z. All buffers and mobile phases for LC separation were prepared in LC-MS grade water purchased from Honeywell (Morris Plains, NJ, USA) and Fisher Scientific (Pardubice, Czechia). All additives added to the LC mobile phases were LC-MS grade.
2.21. MS data processing and statistical analysis
Survey MS and MS/MS spectra were processed in the MaxQuant 1.6.1.0 [57]. Enzyme specificity was set to trypsin/P, and a maximum of two missed cleavages were allowed. Protein N-terminal acetylation, methionine oxidation, glutamine/asparagine deamidation, and N-terminal glutamate to pyroglutamate conversion were selected as variable modifications based on pre-analysis by Preview (Protein Metrics, Cupertino, CA, USA). The derived peak list was searched using the built-in Andromeda search engine in MaxQuant against human reference proteome downloaded in 11th October 2018 including contaminants from UniProtKB database. Workflow used for the determination of redox cysteine changes considered heavy arginine (13C615N4) and lysine (13C6) as variable modifications. Specified iodoTMT labeling has been set as a quantification method. Remaining Group-specific parameters were kept at default values. The minimum Andromeda score needed for modified peptides was set to 0. The minimum ratio count for label-based quantification was set to two quantified peptide pairs. Only unique or razor peptides were considered for calculating protein ratios. For the proteome centric workflow, heavy arginine (13C615N4) and lysine (13C6) were set to Standard type in Group-specific parameters as heavy labels, and specific iodoTMT labeling was added as a fixed modification. The rest of parameters was set as described above. For the analysis of differently oxidized cysteine peptides shown in Fig. 2C, iodoTMT labeling, cysteine dioxidation (-SO2H), and cysteine trioxidation (-SO3H) were set as variable modifications.
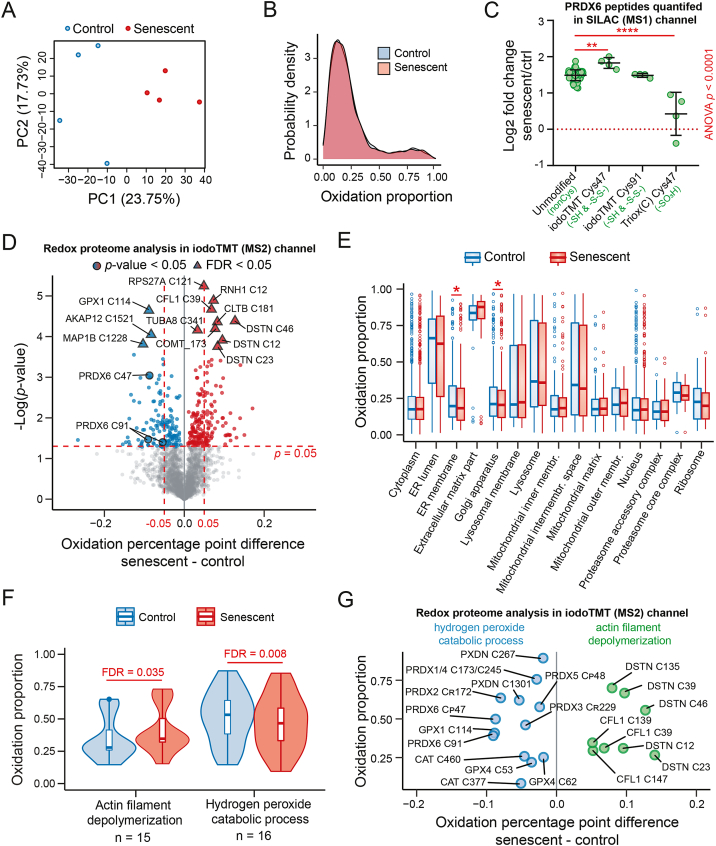
Fig. 2.
High-throughput comparative analysis of the cysteine oxidation status of proliferating and senescent hTERT-RPE-1 cells using SILAC-iodoTMT. (A) Principal component analysis of the oxidation proportion values (n = 2595) quantified in all samples; the percentage indicates proportion of variability explained by principal components 1 (PC1) and 2 (PC2). (B) Distributions of the average oxidation proportion values of control and senescent cells depicted as a density plot. (C) Relative comparison of PRDX6 peptides with different modifications (iodoTMT-labeled or post-translationally modified by oxidation) between senescent and proliferating cells using the SILAC channel. Each dot represents a ratio from one biological replicate. Statistical analysis was performed using one-way ANOVA; pairwise comparisons relative to unmodified peptides were performed using Dunnett's test. Asterisks indicate statistical significance; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (D) The volcano plot shows statistically significant differences between senescent and proliferating cells at the p-value (blue and red circles; two-sample t-test) and FDR level (blue and red triangles; Benjamini-Hochberg correction). (E-F) The cysteine residues of proteins were mapped to cellular compartments (GOCC; E) and biological processes (GOBP; F), and paired samples Wilcoxon test was performed with Benjamini-Hochberg correction. The red asterisk indicates Benjamini-Hochberg FDR < 0.05. The most representative examples were selected for visualization. (G) Selected examples of cysteine residues mapped to the two biological processes shown in (F). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Output files from MaxQuant were processed in R (version 4.0.0) [58] and using Perseus (version 1.6.7.0) [59]. Potential contaminants and proteins identified by site and by reverse sequence were removed. Normalized SILAC H2O2/Ctrl ratios were log2 transformed, and the median value was subtracted to center the median ratio to zero. Only protein groups quantified in all four replicates were selected. Statistical analysis to identify differentially expressed proteins was performed using non-parametric Global Mean Rank test at FDR <0.01 [60]. Fisher's exact test and 1D and 2D Enrichment analysis [61] were performed in Perseus using DAVID annotations [62].
For redox cysteine status quantification, corrected reporter ion intensities for both unlabeled and heavy-labeled cysteine-containing peptides were used to calculate their relative oxidation status as reporter ion intensity oxidized/(reporter ion intensity oxidized + reporter ion intensity reduced) as described previously [48,63]. Only those peptides with quantification values in all four replicates were selected. Batch effects were removed using “ComBat” function from the “sva” R package [64], and the data were normalized using LOESS normalization from “limma” R package [65] prior to statistical analysis (Student's t-test p-value < 0.05 or Benjamini-Hochberg FDR <0.05 as indicated). Statistical analysis to identify differentially oxidized compartments (GOCC) and biological processes (GOBP) was performed using paired Wilcoxon Mann-Whitney test in R (“wilcox.test”) with Benjamini-Hochberg correction. The DAVID bioinformatics resource v6.8 (https://david.ncifcrf.gov) was used to extract the protein annotations (DAVID GOCC DIRECT and DAVID GOBP DIRECT) [62].
2.22. Secretome analysis of senescent cells after PRDX6 silencing
The cells were seeded at a density of 20,000/cm2 and irradiated (20 Gy) 16 h after seeding. The cells were first transfected 2 days after irradiation; the second transfection was performed 5 days after irradiation. Seven days after irradiation, the cells were washed 6 times with PBS, and the cell culture media replaced by serum-free media. The cell culture supernatant was collected after 48 h, i.e., 9 days after irradiation. To remove dead cells and debris, the conditioned media were first centrifuged (200×g, 10 min, 4 °C) and then filter-sterilized using a 0.22 μm syringe filter. Protein concentration was performed using Amicon® Ultra-4 Centrifugal Filter Unit (#UFC801096, Merck Millipore) according to the manufacturer's protocol, the protein concentrate was washed three times with 4 mL of 8 M urea/50 mM Tris-Cl (pH 8.0), and further centrifuged to reach the volume of 60–100 μL. For protein digestion, 20 μg of proteins in 8 M urea/50 mM Tris-Cl (pH 8.0) buffer were reduced using 10 mM TCEP (1000 rpm, 1 h, RT), cysteines were alkylated with 20 mM MMTS (1000 rpm, 30 min, RT, no light exposure), the samples were diluted with 50 mM Tris-Cl (pH 8.0) to reach the final urea concentration of 2 M, and protein digestion was performed using trypsin at 1 : 20 enzyme-to-protein ratio overnight (300 rpm, RT). Peptide samples were acidified using TFA (f.c. = 0.5%) and desalted using Harvard Apparatus C18 Ultra Micro Spin Columns preconditioned by methanol and equilibrated using 0.1% TFA in 2% acetonitrile. Peptides were eluted by 0.1% TFA in 50% ACN and evaporated to dryness.
Samples were re-dissolved and injected into an LC-MS/MS system. The UltiMate 3000 RSLCnano system was used for the liquid chromatography separation. The analytical system consisted of Acclaim PepMap 300, C18, 5 μm, 300 Å Wide Pore, 300 μm × 5 mm trap column and EASY-Spray column, 50 cm × 75 μm ID, PepMap C18, 2 μm particles, 100 Å pore size analytical column. The samples were loaded onto the trap column in 0.05% TFA in 2% acetonitrile at 17.5 μL/min for 3 min. Peptides were separated by a gradient running from 3% to 35% of 80% ACN with 0.1% FA for 90 min at a flow rate of 300 nL/min. The eluted peptides were electrosprayed into Explores 480 using an EASY-Spray ion source (all from Thermo Fisher Scientific, Bremen, Germany).
For library generation, positive ion full scan MS spectra were acquired in the range of 350–1500 m/z using normalized AGC target of 300% and 120,000 resolution with a maximum ion injection time of 100 ms. For MS2, monoisotopic peptide precursor charges 2–6 were selected using an isolation window of 1.6 Da and 60 s of dynamic exclusion. MS2 scans were acquired using NCE 30, 15,000 resolution, normalized AGC target of 75%, and maximum injection time of 75 ms. The instrument was run in a top speed mode with 2 s cycles [66]. DIA data acquisition was performed as described previously [67]. Positive ion full scan MS spectra were acquired in the range of 350–1400 m/z using normalized AGC target of 300% and 120,000 resolution with a maximum ion injection time of 45 ms. AGC target value for fragment spectra was set at 1000%. Forty nine windows of 13.7 Da were used with an overlap of 1 Da. Resolution was set to 15,000, a maximum injection time to 22 ms, and NCE was set to 27.
DIA data analysis was performed in Spectronaut (v14). All DDA and DIA files were used for library generation using the default settings with minor modifications (dithiomethylthiolation of cysteine residues was selected as fixed modification). The targeted data extraction was performed using the default BGS Factory Settings. Both precursor and protein FDR were controlled at 1%. For quantification, “Qvalue” workflow was used for filtering; mean precursor quantity was used for peptide quantification (TOP 3). Cross Run Normalization was performed using “Global Normalization” on “Median” normalization strategy. Interference correction was enabled with minimum 3 MS2 precursors to keep. Other parameters were kept as default unless specified. Pivot protein groups report was exported from Spectronaut and further normalized using LOESS normalization [65] within the NormalyzerDE package [68]. Statistical analysis was performed using Perseus (v1.6.14.0) [59]. Briefly, three siNC-treated samples and four siPRDX6-treated samples (two samples from each siRNA) were grouped, and the data matrix was filtered to only contain protein groups quantified in all samples within at least one group. Missing values were replaced from normal distribution (width 0.3 × standard deviation, downshift 1.8 × standard deviation of the original distribution). Permutation-based FDR was used to identify differentially secreted proteins (FDR <0.05, S0 = 0.4, both sides).
2.23. Data visualization
All boxplots, violin plots, scatter plots and bubble plots were generated using R package “ggplot 2”. In the boxplots, the bold line indicates the median value; box borders represent the 25th and 75th percentiles, and whiskers and the gray panel represent the minimum and maximum value within 1.5 times of interquartile range. Outliers are depicted using solid dots. In the violin plots, the boxplots are combined with kernel density as the violin curve to show the distribution of the data. The white dot marks the median. In the bubble plots, the size of the bubble corresponds to the number of proteins comprised in the category; the color code corresponds to the enrichment factor (Fig. 3C) or the enrichment score (Fig. 5D) calculated in Perseus. The colored scatterplots from blue-to-yellow in Supplementary figures were visualized using R package “LSD”. All grouped scatter plots were created in GraphPad Prism® v8. Prism or R [58] (4.0.0) were used to perform statistical analysis shown in the figures.
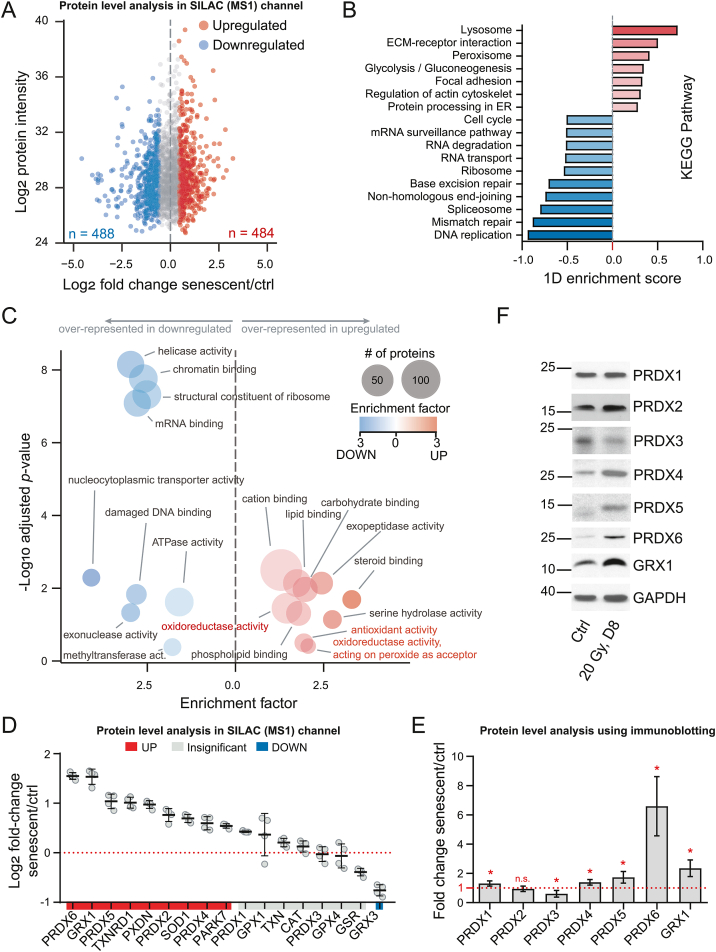
Fig. 3.
Proteomic characterization of the senescent phenotype using SILAC-iodoTMT reveals marked upregulation of antioxidant proteins. (A) Differential protein expression between senescent and control cells; statistical significance was estimated using global mean rank test. Significant proteins (FDR < 0.01) are indicated with blue and red color. (B) KEGG Pathways enrichment analysis was performed using an 1D enrichment test in Perseus. Selected pathways with Benjamini-Hochberg FDR < 0.05 are shown; the length and color of the bar correspond to the enrichment score. (C) Over-representation analysis of up- (right) and down- (left) regulated proteins in cellular senescence. Proteins were annotated with gene ontology molecular function (GOMF) terms, and the p-value (y-axis) was estimated using Fisher's exact test. The size of the bubbles corresponds to number of proteins in the category. (D) Relative abundance of selected antioxidant proteins (corresponding to categories indicated in red in C). The black lines indicate mean ± standard deviation. (E-F) Validation of selected proteins using immunoblotting. At least three independent experiments were performed. GAPDH was used as a loading control. P-values were estimated using one-sample t-test (compared to 1). Asterisks indicate statistical significance; *p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
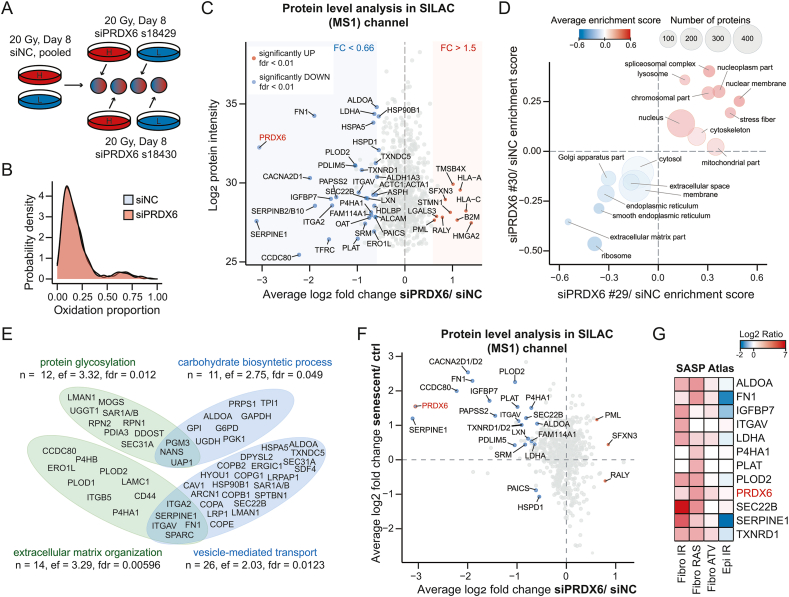
Fig. 5.
Redox-proteomic characterization of the PRDX6 knock-down-induced phenotype in senescent hTERT-RPE-1 cells. (A) Design of a SILAC-iodoTMT experiment with PRDX6 knock-down. (B) Distributions of the average oxidation proportion values of siNC- and siPRDX6-treated cells. (C) Differential protein expression changes induced by siPRDX6 in senescent cells. Significant proteins (a global mean rank test) are shown as indicated; FC = fold change. (D) The two-dimensional enrichment analysis bubble chart of selected cellular compartments. X-axis and Y-axis represent the enrichment scores of siPRDX6 #29 (s18429)/siNC and siPRDX6 #30 (s18430)/siNC ratios, respectively. The size of the bubbles corresponds to number of proteins within each category; the color code corresponds to average enrichment score. (E) Selected categories (GOBP) significantly over-represented in proteins downregulated by siPRDX6 (FDR < 0.05). The Venn diagrams show unique and overlapping proteins included in the categories. Statistical analysis was performed using Fisher's exact test; ef = enrichment factor. (F) Overlap of protein level changes between the senescent/control and the siPRDX6/siNC data set. Indicated proteins were significant in both comparisons (global mean rank test FDR < 0.01). (G) Proteins downregulated by siPRDX6 in senescent cells were mapped to SASP Atlas. The heatmap shows log ratio of the protein secreted by senescent versus control cells in different models of cell senescence as retrieved from SASP Atlas. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Results
3.1. Characterization of the senescence model used for the redox-proteomic analysis
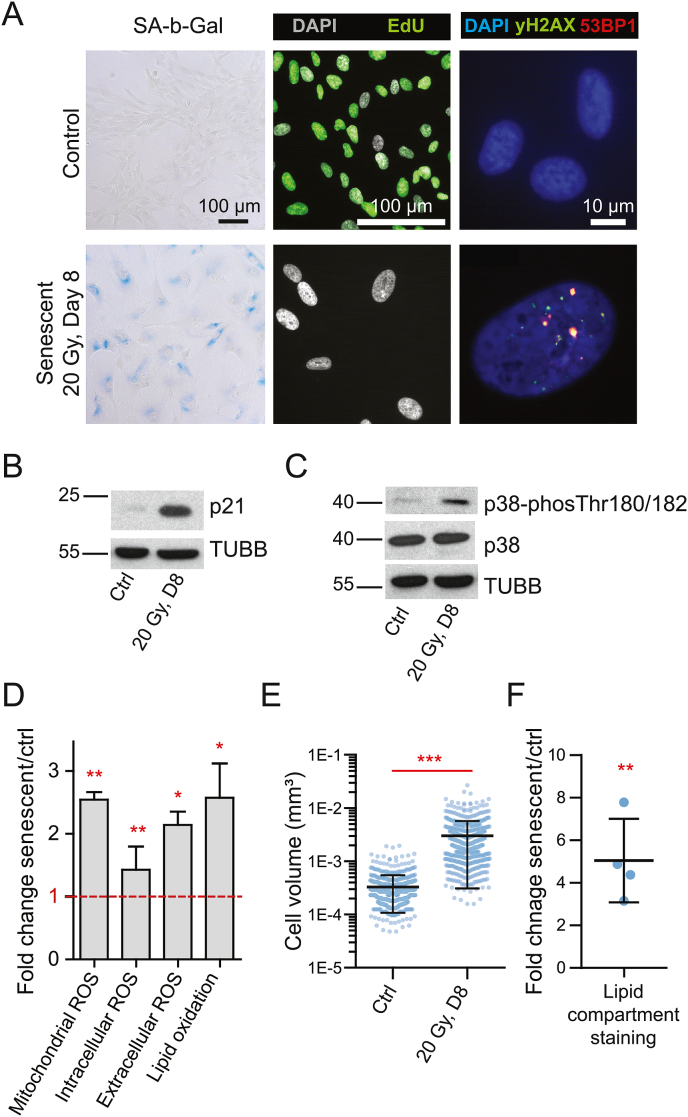
To characterize our model of IRIS, we assessed several well-established markers of cellular senescence in irradiated hTERT-RPE-1 cells (20 Gy; 8 days after irradiation) and compared them to proliferating control cells. The radiation dose of 20 Gy was selected based on our previous experiments [51] to induce a robust and stable cell cycle arrest in this cell line. As depicted in Fig. 1A, the irradiated cells show a typical increase in senescence-associated beta-galactosidase activity, lack of cell proliferation/decreased EdU incorporation (93.2% and 7.5% of positive cells in proliferating and senescent cells, respectively), and persistent DNA damage detected as the presence of 53BP1 and γH2AX foci [69]. Furthermore, irradiated hTERT-RPE-1 cells showed an increased protein level of the cell cycle inhibitor p21 (Fig. 1B). Thus, the presence of several senescence-associated markers confirmed the presence of senescence features in irradiated hTERT-RPE-1 cells.
Fig. 1.
Irradiated hTERT-RPE-1 cells exhibit typical markers of cellular senescence accompanied by increased reactive oxygen species (ROS) levels and oxidative stress response. (A) Cells were irradiated by a dose of 20 Gy, and the senescence-associated beta-galactosidase activity (left panel), EdU incorporation (middle panel), and DNA damage (right panel) 8 days after irradiation were compared to proliferating control cells. (B) Elevated protein level of the cell cycle inhibitor p21 in irradiated and proliferating control cells. β-tubulin (TUBB) was used as a loading control. (C) Increased phosphorylation of p38MAP kinase (p38) and its protein level in irradiated and control cells. β-tubulin (TUBB) was used as a loading control. (D) ROS levels were evaluated 8 days after irradiation by flow cytometry (MitoSOX™ Red Mitochondrial Superoxide Indicator, CellROX™ Deep Red, and BODIPY™ C11, see Methods) to quantify intracellular ROS and Amplex™ Red Hydrogen Peroxide/Peroxidase Assay to quantify extracellular H2O2 production. Fold changes relative to proliferating control cells are shown and were evaluated using one-sample t-test. The bar graphs show mean ± standard deviation from at least 3 biological replicates. (E) Volume estimation of irradiated and proliferating control cells; the scatter dot plots summarize all measurements from three independent replicates, and the black lines indicate mean ± standard deviation. P-value was estimated using Mann-Whitney test. (F) The lipid membrane compartment content was estimated using the BODIPY™ C11 incorporation. Fold changes between senescent and control cells are shown; the black lines indicate mean ± standard deviation, and the p-value was estimated using one-sample t-test (compared to 1). (D-F) The asterisks indicate statistical significance; *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Since the main focus of our study was to analyze the contribution of redox-related changes to development of the senescent phenotype, we further analyzed intra- and extra-cellular ROS production (Fig. 1D). These analyses showed a statistically significant increase in intracellular ROS production (CellROX assay; p = 0.022), mitochondrial superoxide production (MitoSOX assay; p = 0.020), lipid peroxidation (C11 BODIPY assay, oxidized channel; p = 0.0104), and extracellular H2O2 production (Amplex Red assay; p = 0.0116). In general, the interpretation of these results is rather complicated as the irradiated cells have a larger surface, volume (Fig. 1E), and lipid-containing compartments (Fig. 1F). However, phosphorylation of p38α MAP kinase (Fig. 1C), one of the oxidative stress-activated kinases [70], indicates the increased ROS production triggers chronic oxidative stress response in senescent cells. In conclusion, we confirmed the pro-oxidant phenotype in our IRIS model, and therefore, decided to use this model to measure redox proteome changes in cellular senescence.
3.2. Senescence in hTERT-RPE-1 cells is not accompanied by the bulk protein thiol oxidation increase
To simultaneously quantify alterations in protein abundance and cysteine-containing peptide oxidation/reduction in senescent hTERT-RPE-1 cells (20 Gy; day 8), we employed a SILAC-iodoTMT labeling approach combined with LC-MS/MS, as described in our previous study [48]. In this analysis, peptide SILAC precursor ratios (MS1) are used to estimate global changes in peptide and protein level whereas MS/MS fragmentation spectra (MS2) of cysteine-containing peptides contain signals of iodoTMT reporter ions proportional to the fraction of reduced/oxidized sulfhydryl (–SH) groups (Supplementary Fig. 1A). Importantly, the MS1 and MS2 quantitative signals can be used to monitor different cysteine oxidation states. As illustrated in Supplementary Fig. 1A, the iodoTMT (MS2) channel can provide quantitative information about the reduced cysteines (–SH) and reversibly oxidized cysteines (i.e., reducible by TCEP; –S–S– or –SOH), and the data can be used to estimate the proportion of the oxidized cysteines from the total reducible cysteine pool. In addition to this information, the irreversible cysteine oxidation modifications (i.e., not reducible by TCEP; –SO2H and –SO3H) can be detected as modified peptidoforms in the MS1 and quantified using SILAC. Thus, monitoring the irreversible oxidation using SILAC can diminish the limitation of the iodoTMT labeling in addressing the quantity of higher oxidation states [48].
SILAC-iodoTMT experiment presented in this study was conducted in two label-swap arrangements. In the first arrangement, the SILAC “heavy” channel was used to label the senescent group, while in the other one the “heavy” channel was used to label proliferating cells. Both arrangements were performed in two biological replicates for proliferating control and senescent cells (Supplementary Fig. 1A). In total, we retrieved quantitative information about the redox status (proportion of reversibly oxidized cysteine from the total signal, i.e., ox/(ox + red)) of 17,704 cysteine residues; 2595 of them were quantified in both SILAC light and heavy channels and all four biological replicates and were considered for downstream analysis. Although the senescent cells were clearly separated from the proliferating counterparts in the principle component analysis (PCA; Fig. 2A), we did not detect any global difference in the cysteine oxidation status between senescent and proliferating cells (Fig. 2B), and the values correlated strongly between the groups (rho = 0.96; Supplementary Fig. 1B). However, we found a number of differentially oxidized cysteine residues between the groups (Fig. 2D and Supplementary Table 1). Further analysis showed the distinct oxidation status of different compartments and also sub-compartments (Fig. 2E; Supplementary Table 2), which was in concordance with known subcellular compartmentalization of redox potentials in cells [71]. Moreover, there were rather small, but significant differences in cysteine oxidation on the level of individual organelles between senescent and proliferating cells, for example in ER membrane- and Golgi apparatus-annotated proteins (Fig. 2E; Supplementary Table 2). Interestingly, we also observed a non-significant trend for proteins of the extracellular matrix (“extracellular matrix part”), which appeared to be more oxidized in senescent cells potentially correlating with the observed increase of extracellular ROS production shown in Fig. 1D. Furthermore, a similar analysis of gene ontology biological process terms (GOBP) revealed several processes that were associated with increased oxidation of cysteine residues in senescent cells (Fig. 2F; Supplementary Table 2), such as “actin filament depolymerization”, and on the other hand, less oxidized, such as “hydrogen peroxide catabolic process”. The first group contains redox-regulated cysteines from the actin-associated cytoskeletal proteins cofilin and destrin controlling actin dynamics (reviewed in Ref. [72]) suggesting redox-dependent control of cytoskeleton in senescent cells. The latter group corresponds to proteins from the Prx family and other antioxidant proteins (e.g., CAT, GPX1, GPX4, PDXN, PRDX1, PRDX2; Fig. 2G) including important catalytic cysteines such as resolving Cys172 of PRDX2, resolving Cys229 of PRDX3, peroxidatic Cys48 of PRDX5, and peroxidatic Cys47 of PRDX6. Thus, this analysis revealed a striking and unexpected observation that in senescent cells these redox-sensitive cysteine residues might be less oxidized despite the increased ROS production in senescent cells.
As shown in Fig. 2D, peroxiredoxin 6 (PRDX6) Cys47 and Cys91 residues appeared to be significantly less oxidized in senescent cells, an unexpected observation identical to our recent study in H2O2-treated cells [48]. In this study, we proposed that due to the Cys47 hyperoxidation, a part of the cysteine pool could not be reduced by TCEP during the iodoTMT labeling and resulted in underestimation of the oxidized proportion of this cysteine. This was suggested by a relatively lower level of reducible Cys47 PRDX6 peptide in H2O2-treated cells. We thus explored whether we could observe the same Cys47 behavior in IRIS. To do so, we performed an orthogonal data analysis to analyze different cysteine oxidation states using the SILAC MS1 channel. We estimated the fold changes (senescent/control) of the iodoTMT labeled (i.e., sum of the reduced and TCEP-reducible oxidized form, as the two forms can be distinguished only in MS2) or hyperoxidized (Triox(C); not reducible by TCEP) cysteinylated peptides and compared their SILAC ratios to the unmodified PRDX6 peptide pool (Fig. 2C and Supplementary Fig. 14). The dioxidized (–SO2H) version of PRDX6 Cys47 was not detected. By doing so, we revealed that the ratios of peptides bearing different modifications showed significantly different fold change distributions (ANOVA p < 0.0001) indicating they exist with a different stoichiometry in senescent and proliferating cells. Strikingly, the fold change of the hyperoxidized form of Cys47 (i.e., sulfonic acid, –SO3H residue) was significantly lower compared to unmodified peptides (Fig. 2C and Supplementary Fig. 14B). This indicated that there are more molecules of the hyperoxidized Cys47 PRDX6 proteoform in senescent cells, but they represented a smaller proportion of all PRDX6 molecules compared to proliferating control. Complementary to this observation, the fold change of the iodoTMT-labeled form of peroxidatic Cys47 was greater than the ratio of unmodified PRDX6 peptides, indicating that more reduced/TCEP-reducible Cys47 peptidoforms are present in senescent cells (Fig. 2C and Supplementary Fig. 14B). These results suggested that proportionally, in senescent cells, Cys47 PRDX6 exists in the hyperoxidized form with a lower stoichiometry than in proliferating cells, despite more ROS generated in IRIS. Furthermore, this analysis highlighted different regulation of PRDX6 Cys47 under conditions of acute oxidative stress induced by H2O2 treatment [48] and chronic oxidative stress in IRIS.
We further applied the same analysis to examine whether the iodoTMT quantification of the unexpectedly less oxidized cysteine residues corresponding to hydrogen peroxide metabolism and presented in Fig. 2G could reveal similar conclusions. In contrast to PRDX6, we did not detect any higher oxidation forms of the cysteine residues shown in Fig. 2G suggesting that if they are present in the cells, they are most likely present with a low stoichiometry (Supplementary Fig. 13). However, similarly to our previous study [48], for most of the cysteine residues, we could compare the SILAC ratios of the iodoTMT-labeled peptides with the ratios of the unmodified peptides pool. Importantly, for most of the presented cysteines, the ratio of the iodoTMT-labeled peptide was not significantly lower diminishing concerns these residues were blocked by a non-reducible modification. However, we found a significantly lower ratio of the mitochondrial PRDX3 Cys229 suggesting this cysteine could have been modified by an unknown non-reducible modification (Supplementary Fig. 13).
In conclusion, the redox-proteomic analysis suggested the increase of ROS in IRIS was not accompanied by bulk increase of cysteine thiol oxidation. Nevertheless, significant differences on the level of individual proteins and cysteine residues, compartments, and biological processes suggested the redox status of senescent cells was altered, potentially contributing to shaping the senescent phenotype.
3.3. Proteomic analysis of senescent cells reveals upregulation of antioxidant proteins in IRIS
In addition to the cysteine redox status, the SILAC-iodoTMT analysis provided information about senescence-associated proteome changes quantified in the SILAC channel. In total, 4414 protein groups were identified; 1995 of them were quantified in all four biological replicates, among them 484 were upregulated and 488 downregulated (FDR < 0.01; Fig. 3A and Supplementary Table 3). Pathway analysis (KEGG) revealed typical pathways up- and downregulated in senescent cells (Fig. 3B; Supplementary Table 4), such as enrichment of “lysosome” and “ECM-receptor interaction” in the upregulated proteins and enrichment of “DNA replication” and “spliceosome” within the downregulated proteins. In addition to the senescent markers shown in Fig. 1, this analysis further confirms irradiated hTERT-RPE1 cells developed typical features of the senescent phenotype.
The enrichment analysis of gene ontology molecular function terms (GOMF) revealed several categories significantly enriched in the fraction of proteins upregulated in irradiated cells related to antioxidant functions such as “oxidoreductase activity” and “antioxidant activity” (Fig. 3C; Supplementary Table 5), suggesting the antioxidant proteins were upregulated after irradiation to potentially compensate for the increased ROS production. The relative level of proteins included in these categories is shown in Fig. 3D. Several members of the PRDX family were upregulated in senescence, namely PRDX6, PRDX5, PRDX4, or PRDX2 together with members of the thioredoxin and glutaredoxin systems, glutaredoxin 1 (GRX1) and thioredoxin-1-reductase (TXNRD1), and other antioxidant proteins, e.g., superoxide dismutase 1 (SOD1) and Parkinson disease protein 7 (PARK7). We also confirmed the increased level of selected proteins from this group (GRX1, PRDX6, PRDX4, and PRDX5) by immunoblotting (Fig. 3E and F). Moreover, PRDX4, PRDX5, and PRDX6 were also significantly induced at the transcript level (Supplementary Fig. 2A). Thus, irradiated hTERT-RPE-1 cells increased both the transcript level and the level of central proteins involved in ROS metabolism and maintenance of the protein thiol redox state, possibly to compensate oxidative stress in senescence and to prevent the dysregulation of redox homeostasis and accumulation of protein oxidation damage.
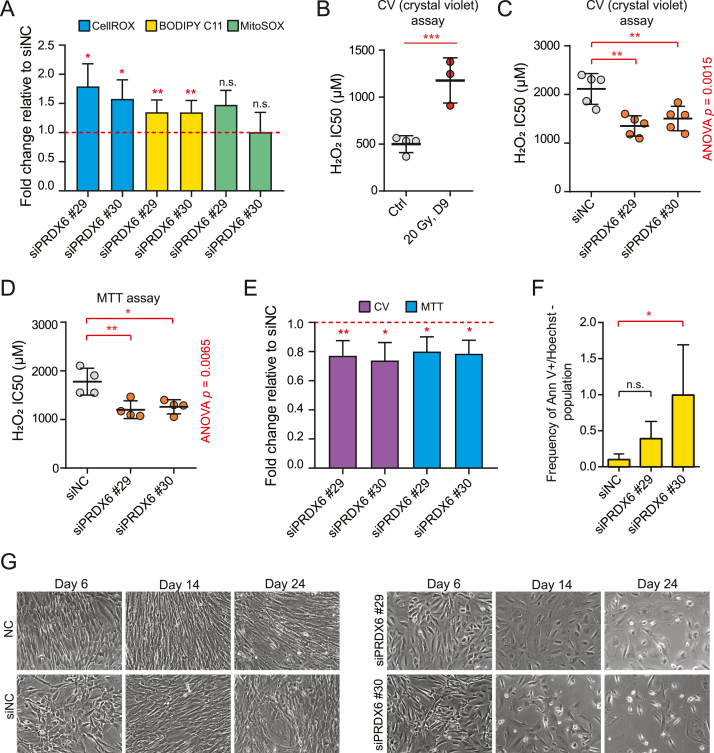
3.4. PRDX6 silencing increases intracellular ROS levels, sensitizes senescent cells to hydrogen peroxide treatment, and induces death of senescent cells
To investigate whether the upregulation of antioxidant proteins contributed to the buffering of ROS-induced redox changes in senescent cells, we performed a systematic silencing of selected genes using specific siRNAs and measured the effect of the silencing on intracellular ROS. The effectiveness of RNA interference was first validated using RT-qPCR and immunoblotting (Supplementary Fig. 2B – D). Specifically, based on our proteomic results and immunoblotting validation, we performed silencing of the following four genes: GRX1, PRDX4, PRDX5, and PRDX6. Among these targets, only the silencing of PRDX6 induced a significant increase of ROS levels in senescent cells (Fig. 4A and Supplementary Fig. 3A). As the PRDX6 peroxidase activity also encompasses phospholipid hydroperoxides as substrates, PRDX6 plays a role in the repair of peroxidized cell membranes [73]. Therefore, we assessed lipid peroxidation upon PRDX6 downregulation in senescent cells. Indeed, the knock-down significantly enhanced the proportion of oxidized lipid membrane compartments (Fig. 4A). On the other hand, siPRDX6 affected mitochondrial ROS inconsistently (Fig. 4A) and did not impact extracellular ROS production (Supplementary Fig. 3B). Thus, in irradiated hTERT-RPE-1 cells, the PRDX6 activity significantly contributes to the protection against excessive intracellular ROS and lipid peroxidation.
Fig. 4.
PRDX6 knock-down mediates increase in ROS levels and cell death of senescent hTERT-RPE-1 cells. (A) ROS levels were evaluated 8 days after irradiation by flow cytometry using CellROX™ Deep Red (total ROS, n = 4), BODIPY™ C11 (lipid peroxidation, oxidized/total, n = 6), and MitoSOX™ (mitochondrial superoxide, n = 3) staining to quantify intracellular ROS in senescent cells transfected with siPRDX6 s18029 (#29) or s18030 (#30). Fold changes (FC) relative to irradiated cells transfected with siNC are shown. (B) H2O2 IC50 was estimated using crystal violet staining in control (n = 4) and senescent (n = 3) cells; p-value was estimated using a two-sample t-test. (C-D) H2O2 IC50 was estimated using crystal violet staining (C; n = 5) and the MTT assay (D; n = 4) in senescent cells after PRD6 knock-down. Statistical analysis was performed using ANOVA with Dunnett's multiple comparisons test. (E) Viability of senescent cells transfected with siPRDX6 s18029 (#29) or s18030 (#30) was analyzed relative to cells transfected with siNC using crystal violet staining (n = 5) and the MTT assay (n = 4). (F) The proportion of annexin V-positive and Hoechst 33258-negative cells was evaluated using flow cytometry (n = 3). Statistical analysis was performed using Friedman test with Dunn's multiple comparison test. (G) Loss of the adherent fraction of cells after repeated knock-down of PRDX6 in irradiated cells. Representative pictures for selected time points are presented. (A-F) In all bar graphs, mean ± standard deviation are shown. In all scatter plots, the lines indicate mean ± standard deviation; each dot corresponds to an independent biological replicate. Statistical analysis of fold changes relative to siNC was performed using a two-sided one-sample t-test (compared to 1). The asterisks indicate statistical significance; *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Senescent cells, in dependence on the cell type and the model of cellular senescence, typically show an increased resistance to cell death. Inspired by the observation the antioxidant proteins are markedly upregulated in our model of senescence, we aimed to investigate the sensitivity of irradiated cells to H2O2 as compared to proliferating control cells. Indeed, the H2O2 sensitivity of senescent cells was lower than of proliferating cells (Fig. 4B), and the IC50 positively correlated with the time point after irradiation (2, 6, 9, 12, and 16 days after IR; Supplementary Figs. 4A and 4B) indicating this effect cannot be explained simply by the difference between proliferating and nonproliferating cells. In addition to H2O2, the sensitivity to another oxidation agent, diamide, also decreased over time (Supplementary Fig. 4C). As some of the antioxidant proteins were observed to be rising over all time points in a label-free proteomic analysis (Supplementary Fig. 4D), the increase of resistance to H2O2 expressed as IC50 strongly correlated with the upregulation of these proteins (Supplementary Fig. 4E). Thus, we also investigated whether the H2O2 resistance could be attenuated by silencing of the four selected antioxidant genes. In concordance with the significant effect on ROS production, only PRDX6 silencing unequivocally sensitized senescent cells to oxidative stress (Fig. 4C and D and Supplementary Figs. 5A–F).
Furthermore, siPRDX6 itself moderately decreased the viability of senescent cells in crystal violet staining and MTT assays (Fig. 4E), while silencing of GRX1, PRDX4, or PRDX5 did not (Supplementary Fig. 6A), and slightly increased the number of annexin V-positive apoptotic cells in senescent cell culture (Fig. 4F and Supplementary Fig. 6B). To further examine the phenotype induced by downregulation of PRDX6, we performed several re-transfections of proliferating and senescent cells to monitor the long-term effects of siPRDX6. Strikingly, we observed slow but continuous loss of adherent cells in siPRDX6-treated senescent cell culture over time (Fig. 4G and Supplementary Figs. 7A and 7B). Furthermore, this effect was not observed in proliferating cells, in which only siPRDX6 s18029 caused slower proliferation, but no cell death was observed over the time course of the experiment (Supplementary Fig. 6C).
In conclusion, senescent cells progressively increase the resistance to oxidative stress by augmentation of the antioxidant system. Furthermore, systematic silencing of antioxidant genes elevated in response to irradiation suggested PRDX6 expression is the key factor maintaining senescent cell viability in conditions of increased oxidative stress and provides an advantage against extracellular oxidative stress induced by the H2O2 treatment.
3.5. PRDX6 silencing suppresses senescence-associated upregulation of extracellular matrix proteins
As PRDX6 silencing led to increased ROS levels in senescent cells accompanied by noticeable cell death and sensitization to H2O2, we hypothesized that PRXD6 downregulation might impair antioxidant capacity of senescent cells resulting in redox homeostasis disruption as has been shown by studies in proliferating cells [32,33]. Thus, we applied our SILAC-iodoTMT method to describe differences between senescent cells transfected by siNC and siPRDX6 (s18029, s18030) in biological duplicates and using swapped SILAC “light” and “heavy” channels (four replicates in total; see Fig. 5A and Supplementary Fig. 8A). Although the conditions were clearly separated in PCA analysis (Supplementary Fig. 8B), and several cysteine residues showed statistically significant differences in the oxidation level upon siPRDX6 (Supplementary Fig. 8C) indicating PRDX6 knock-down indeed affected redox homeostasis, we did not observe any global dysregulation of cysteine oxidation (Fig. 5B).
Next, we investigated protein level changes in the SILAC channel to provide insight into the H2O2-sensitive phenotype described above. As illustrated by Fig. 5C, PRDX6 silencing induced significant differences (FDR < 0.05) in the relative abundance of 186 proteins as compared to senescent cells treated with siNC only (Supplementary Table 6). The majority of them were downregulated (n = 103) with PRDX6 being the second most downregulated protein, further confirming the efficiency of the knock-down (Fig. 5C). To further describe the siPRDX6-induced phenotype and inspect the concurrence between two different siRNAs, we performed a 2D-enrichment analysis. Fig. 5D depicts cellular compartments up- and downregulated following the PRDX6 silencing (2D enrichment analysis FDR < 0.05; Supplementary Table 8). The effect of both PRDX6 siRNAs on the major compartments was consistent except for mitochondria. Interestingly, we detected a significant downregulation of extracellular matrix proteins, smooth ER, and Golgi apparatus. The effect on cellular compartments was further validated by flow cytometry, which confirmed the decreased signal of the membrane-incorporating Bodipy C11 probe in the siPRDX6-treated cells (Supplementary Fig. 8D) and the inconsistent effect of the siRNAs on mitochondrial staining (Supplementary Fig. 8E). Using an over-representation analysis of the biological processes associated with the downregulated proteins (Fig. 5E and Supplementary Table 7) we identified that the silencing affected metabolism of carbohydrates, protein synthesis, protein glycosylation and transport, and organization of extracellular matrix in senescent cells (Fig. 5E). Altogether, in addition to effects on carbohydrate metabolism reported before [33], these analyses suggested that protein secretion and extracellular matrix composition might be altered by PRDX6 downregulation in senescent cells.
To specifically address which of the downregulated proteins were upregulated in IRIS compared to proliferating control cells (i.e., significantly upregulated in the first SILAC-iodoTMT experiment), we correlated the protein level results from the two experiments (Fig. 5F). As depicted in the upper left quadrant, PRDX6 silencing led to decrease of the relative abundance of some of the most upregulated proteins in senescent hTERT-RPE-1 cells such as SERPINE1, CCDC80, or FN1, which are all components of the extracellular matrix of senescent cells. As extracellular matrix proteins have been described as a component of senescence-associated secretory phenotype (SASP), we asked which of the most significant proteins from this quadrant (significantly upregulated in senescence and significantly downregulated by siPRDX6 in senescence, both at FDR < 0.01; n = 19) have been already described as components of SASP, including PRDX6 itself. Twelve of these proteins were retrieved from the SASP Atlas [74], most of them differentially secreted in at least one condition inducing cellular senescence (Fig. 5G). In conclusion, the results revealed PRDX6 loss affected the metabolism and composition of extracellular matrix in senescent hTERT-RPE-1 cells.
3.6. PRDX6 silencing alters the composition of senescent secretome and suppresses senescence-associated secretion of IL-6
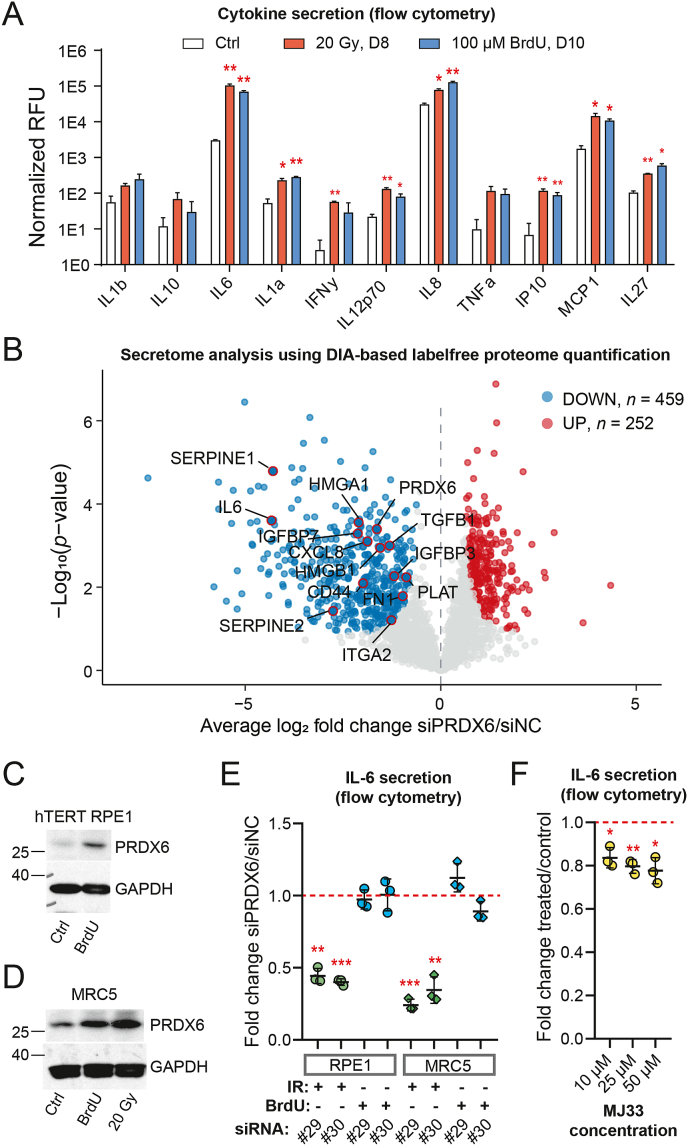
One of the most prominent components of the SASP are pro-inflammatory cytokines and chemokines [75]. Thus, we first investigated whether irradiation and 5-bromo-2′-deoxyuridine (BrdU; another senescence inducer [76]) treatment induced increased secretion of these molecules in hTERT-RPE-1 cells using a multiplexed cytokine array. As shown in Fig. 6A, concentration of a number of known pro-inflammatory factors, including IL-1α, IL6, or IL8, increased in the cell culture media, confirming senescent hTERT-RPE-1 cells develop a pro-inflammatory phenotype. These three cytokines were also differentially expressed at the mRNA level in irradiated hTERT-RPE-1 cells (Supplementary Fig. 9A) suggesting their expression was regulated at the transcriptional level.
Fig. 6.
PRDX6 silencing alters the composition of senescent secretome and suppresses ionizing radiation-induced senescence-associated secretion of IL-6. (A) Relative levels of secreted proteins in cell culture media normalized to cell count per condition; statistical analysis was performed using repeated measures ANOVA with Dunnett's correction. Asterisks indicate significance in comparison to untreated control. RFU – relative fluorescence units. (B) The volcano plot illustrates changes in senescent cell secretome after PRDX6 knock-down measured by label-free DIA LC-MS/MS proteomics. Significant proteins (permutation-based FDR < 0.05, S0 = 0.4) are marked with blue and red colors. (C) PRDX6 protein level in hTERT-RPE-1 cells after treatment with 100 μM BrdU (10 days); GAPDH was used as a loading control. (D) PRDX6 protein level in MRC-5 cells after 100 μM BrdU or 20 Gy of IR (10 days); GAPDH was used as a loading control. (E) Relative levels of IL-6 secreted by senescent hTERT-RPE-1 or MRC-5 cells after 100 μM BrdU or irradiation (20 Gy) and after PRDX6 knock-down by the siRNA indicated; #29 = s18429, #30 = s18430. Fold changes relative to senescent cells transfected with siNC are shown; statistical analysis was performed using one-sample t-test. (F) Relative levels of IL-6 secreted by senescent hTERT-RPE-1 (20 Gy, 9 days) after MJ33 treatment (72 h, concentrations as indicated; inhibitor of iPLA2 activity of PRDX6). Fold changes relative to untreated senescent cells are shown; statistical analysis was performed using one-sample t-test. (A, E, F) The relative concentration of cytokines was estimated using a Human Inflammation 11-Plex Panel kit. Asterisks indicate statistical significance; *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Inspired by our previous proteomics results, we next performed a secretome analysis following PRDX6 knock-down to identify whether it has any significant impact on SASP. Using a label-free DIA approach, we identified more than 700 proteins that were differentially secreted upon siPRDX6, and most of them were significantly downregulated (Fig. 6B and Supplementary Table 9). Further inspection of the data revealed downregulation of many proteins associated with SASP, such as SERPINE1, HMGB1, TGFB1, or IGFBP7. From 459 proteins downregulated after PRDX6 silencing, 216 (47%) were previously reported in the SASP Atlas [74]. Most importantly, the analysis identified a significant change in secretion of the pro-inflammatory cytokine IL-6. Thus, this analysis suggested that cells lacking PRDX6 developed an alternative SASP composition than the control senescent cells.
To confirm and extend our findings to different SIPS models, we analyzed the effect of siPRDX6 in irradiated and BrdU-treated hTERT-RPE-1 and MRC-5 cells. First, we evaluated whether PRDX6 upregulation is a common feature in these models. We confirmed that the stress increased the abundance of PRDX6 at both the protein and mRNA levels in all conditions (Fig. 6C&D and Supplementary Fig. 9B, respectively) and increased the IL-6 mRNA expression (Supplementary Fig. 9B), and we also confirmed siPRDX6 decreased expression of PRDX6 mRNA (Supplementary Fig. 9C). Next, we confirmed that PRDX6 silencing significantly decreased secretion of IL-6 in irradiated hTERT-RPE-1 cells as indicated by the proteomic results (Fig. 6E). Interestingly, the analysis of both IR- and BrdU-induced senescence in two different cell lines showed this effect was not cell line-specific but insult-specific, and was significant in both cell lines upon irradiation (Fig. 6E and Supplementary Figs. 10 and 11).
Since PRDX6 exhibits multiple enzymatic activities including the iPLA2 activity which has been previously shown to be involved in pro-inflammatory pathways [28,35,36], we further asked whether the iPLA2 activity is required for the upregulation of IL-6 in IRIS. To do so, we performed experiments with a specific inhibitor of PRDX6 iPLA2 activity MJ-33 [56]. The results showed that the MJ-33 treatment significantly decreased secretion of IL-6 (Fig. 6F and Supplementary Fig. 12A), although not to the level observed in the silencing experiments. Contrary to the silencing experiments, we did not observe any loss of cell adhesion within 72 h in any concentration used (Supplementary Fig. 12B). Thus, the iPLA2 activity of PRDX6 contributed to IL-6 secretion in IRIS.
To conclude, these experiments provided a striking evidence that PRDX6 suppression affects the development of the senescent phenotype in IRIS. More specifically, IL-6 secretion was observed to be dependent on PRDX6 in IRIS, which is to some extent related to the loss of its iPLA2 activity.
4. Discussion
Constitutive production of high ROS levels associated with the induction and maintenance of the senescent phenotype [77] can alter the function of cellular macromolecules by inducing oxidative modifications of DNA and RNA, lipids, and proteins [78]. Among the variety of protein oxidative modifications, the reversible redox-sensitive cysteine oxidation to sulfenic acid (–SOH) presents an important regulatory switch in response to intracellular redox changes and thus contributing to redox signaling pathways [78]. Redox proteomics is an emerging field facilitating large-scale investigation of redox cysteine signaling in physiological and pathophysiological states [48,[79], [80], [81], [82], [83], [84], [85], [86], [87]]. Although many studies have linked cellular senescence to oxidative stress, no study has attempted to neither describe the redox proteome changes on a large-scale nor to address whether there is a mechanistic role of ROS-induced post-translational modifications of proteins in the development and maintenance of the senescent phenotype.
Here, we applied our recently developed method, SILAC-iodoTMT [48], to simultaneously assess protein abundance and cysteine redox status changes using LC-MS proteomics and to explore the role of ROS in the development of a well-characterized model of radiation-induced premature senescence [51] accompanied by elevated levels of intra- and extra-cellular ROS (Fig. 1). Several reports have suggested a change in bulk cysteine oxidation under various conditions [80,88,89], however, studies investigating the redox proteome during aging in vivo [86,89] have rather identified rewiring or dysfunction of redox signaling networks [86]. Our in vitro study shows that alterations in the redox system in hTERT-RPE-1 cells undergoing IRIS resemble changes observed in in vivo aging models. In contrary to the hypothesis that increased ROS levels would affect the cysteine residues nonspecifically and globally, we observed up- and downregulation of thiol oxidation in groups of proteins with specific biological functions (Fig. 2). On one hand, several redox-sensitive cysteines regulating the cytoskeleton dynamics were more oxidized in senescent cells. On the other hand, and quite unexpectedly, redox-sensitive cysteines of well-known antioxidant proteins including peroxidatic and resolving cysteines from several Prx family members appeared to be less oxidized in senescent cells. Due to the limitation of iodoTMT for labeling higher oxidation forms, we further used SILAC quantification of different peptidoforms to address whether the paradoxical oxidation decrease in these redox-sensitive cysteines was caused by their hyperoxidation resulting in the signal loss as described before [48]. Importantly, this was not the case for most cysteine residues confirming the lower oxidation status of these cysteines (Supplementary Fig. 13). However, we did detect a significant iodoTMT-labeled peptide signal loss for the mitochondrial PRDX3 Cys229, and on the other hand, the analysis of PRDX6 peptides provided a deeper insight into PRDX6 modifications of Cys47 stoichiometry between senescent and proliferating cells. First, these results suggested that even in cells with higher oxidation levels, the redox modifications tend to occur at very specific residues consistently to previous reports [63]. Second, these results also emphasized the limitations of iodoTMT and the importance of parallel monitoring of the MS2 iodoTMT channel and MS1 SILAC channel for peptide quantification to improve data interpretation for individual cysteine residues. For example, in a previous iodoTMT-based redoxome study [63] investigating the response to hydrogen peroxide treatment, the authors reported a marginal reductive shift in most cysteine thiols and identified tens of cysteine residues with significantly decreased oxidation. The peptide-level information determined by SILAC could help to further resolve if these cysteines were less oxidized or potentially modified with a non-reducible modification [48]. Additionally, by simultaneous assessment of protein abundance changes we revealed upregulation of a class of proteins with antioxidant capabilities as a response to the senescence-inducing stimuli (Fig. 3). Thus, when the cells expand their size and volume, these important antioxidant proteins are not diluted in the higher protein content of the senescent cells. On the contrary, some of them relatively increase their level as an adaptive response to the stress triggered by irradiation. Furthermore, the very specific nature of the oxidation changes also supports the idea of an adaptive stress response. Therefore, we concluded that senescent cells augment their antioxidant system to successfully establish a new redox homeostasis in response to long-term oxidative stress.
In our previous study [48], we investigated the short-term effect of hydrogen peroxide treatment, which in long-terms has also been shown to ultimately lead to cellular senescence [3]. We reported an unexpected reduction of several resolving cysteines in the very early phase, after 5 and 60 min post-treatment, and a secondary bulk cysteine oxidation 32 h post-treatment accompanied by increased ROS, increased levels of stress-response proteins, decreased levels of cell cycle proteins, and in general a cell phenotype change correlating with frequently observed senescence-associated changes. However, at this timepoint, we did not observe any significant increase of redox-related proteins. In this study we now add a long-term observation, 8 days post senescence-inducing stimuli, which is accompanied by increased ROS, but contrary to the short-term treatment, no bulk cysteine oxidation and a marked increase of antioxidant proteins were observed. Based on these results we could hypothesize the following dynamics in the redoxome of cells undergoing stress-induced cellular senescence. The stress induces a temporary increase of cysteine oxidation which in turn could contribute to upregulation of the antioxidant system and establishment of a new redox homeostasis which is different from the original state, but the emerging ROS are still compensated enabling maintaining cell function and survival in the conditions of the chronic oxidative stress.
Senescent cells are considered to be less sensitive to apoptosis due to several mechanisms dysregulating the balance between pro- and anti-apoptotic proteins or resistance to extrinsic apoptosis stimuli [[90], [91], [92]], which had led to a proposal of several senolytic drugs targeting the programmed cell death pathways [93,94]. Consistently with previous observations [95,96], senescent hTERT-RPE-1 cells were markedly more resistant to apoptosis induced by H2O2 than proliferating cells in a time-dependent manner correlating with the relative abundance of antioxidant proteins over time. To decipher the individual contribution of four antioxidant proteins revealed by proteomic profiling (GRX1, PRDX4, PRDX5, and PRDX6) to the cellular redox homeostasis and ROS resistance of senescent cells, we performed a series of silencing experiments (Fig. 4). Among these candidate proteins, the silencing of PRDX6, increased intracellular ROS and lipid peroxidation in senescent cells and sensitized them to H2O2-induced cell death. More importantly, we also noticed that a long-term PRDX6 knock-down induced cell death independently on any other stress stimuli, and the detected apoptosis in senescent cells suggesting PRDX6 might play an important role in maintaining their viability. Several reports have already shown that loss of PRDX6 or other peroxiredoxins led to increased sensitivity to oxidative damage-induced cell death (see, for example [19,97]); however, to our best knowledge, no study has indicated that PRDX6 might be indispensable for the cell viability in IRIS in defense to excessive oxidative stress.
To gain more insight into the role of PRDX6 in maintaining the senescent cells redox homeostasis and viability, we performed redox-proteomic profiling after PRDX6 knock-down (Fig. 5). The redox-proteomic analysis has been previously performed in only two studies using PRDX6 knock-down [32] or CRISPR/cas9 knockout [33] in proliferating HepG2 cancer cell line. In both studies, PRDX6 loss led to reduced proliferation, and consistently with our experiments in proliferating cells, no induction of cell death was observed. The studies further suggested differential redox regulation of multiple metabolic pathways in PRDX6-deficient cells. On the contrary, downregulation of PRDX6 in senescent cells did not have any major impact on oxidation of redox-sensitive proteins in metabolic pathways; nevertheless, it must be noted that the limited depth of our redox-proteomic data did not enable to fully compare the data sets together. Another striking difference between the data sets was the behavior of mitochondrial proteome, since we did not detect any downregulation of mitochondrial proteins which was further confirmed by mitochondrial staining intensity. Thus, although several studies highlighted the importance of PRDX6 for mitochondrial health [33,[98], [99], [100]], PRDX6 knock-down did not seem to contribute to further alteration of mitochondrial function in senescent cells. On the other hand, the proteomics profiles revealed some similarities between proliferating and senescent cells lacking PRDX6, such as differential expression of enzymes of the key metabolic pathways. Moreover, we further observed downregulation of components of ribosomes, ER, and Golgi apparatus parts and changes in extracellular matrix composition in PRDX6-deficient senescent cells.
Senescent cells secrete pro-inflammatory factors, collectively known as the senescence-associated secretory phenotype or SASP [75], which in the long term can promote chronic inflammation, aging-associated diseases, and cancer [101,102]. By comparing the proteomic profile of siPRDX6-treated senescent cells to a recently published proteomic SASP Atlas [74], the data suggested SASP modulation in PRDX6 knock-down. To further elaborate on this striking observation, we extended our proteomic analysis to the SASP analysis (Fig. 6). The analysis revealed that protein secretion is indeed altered after PRDX6 knock-down with a strong preference for downregulation. The downregulated proteins comprised a number of key pro-inflammatory factors previously linked to senescence and its pathophysiology such as HMGB1 [103], SERPINE1 and SERPINE2 [104], FN1 [105,106], or PLAU [104], and even more importantly, several cytokines, such as TGFB1, IL-6, or IL-8. As a striking example, we identified and validated the downregulatory effect of PRDX6 knock-down on IL-6 secretion, which was only observed in IRIS, but not BrdU-induced senescence in hTERT-RPE-1 and MRC-5 cell lines (Fig. 6E). Few studies have previously linked cytokine expression to PRDX6. For example, in a mouse model of dextran sodium sulfate (DSS)-induced colitis [107], Prdx6 knock-out mice were shown to be more resistant to DSS-induced colitis and did not upregulate expression of several cytokines including TNF-α, IFN-γ, IL1-β, and IL-6. On the other hand, upon LPS treatment, PRDX6 knock-down cells showed increased NF-κB-dependent expression of pro-inflammatory cytokines such as TNF-α, IL1-α, IL1-β, or IL-8 [108]. Furthermore, PRDX6 overexpression alleviated adriamycine-induced myocardial injury in rats and decreased expression of IL1-β and IL-6 [109]. Collectively, these studies reported rather contradictory findings, suggesting the association of PRDX6 with cytokine expression might be context-dependent and probably related to its different enzymatic activities and cellular localization of PRDX6.
Similarly to other Prx family members, PRDX6 has been previously linked to inflammation in a multi-faceted way [34]. The peroxidase activity of PRDX6 is most often described as anti-inflammatory, preventing oxidation damage, while its iPLA2 has been previously linked to pro-inflammatory phenotypes [28,35,36]. The iPLA2 activity of PRDX6 has been further shown to contribute to lipid signaling by releasing arachidonic acid, which leads to increased proliferation in melanoma [41], invasion and metastasis in lung cancer cells [39], or production of IL-1β in response to proinflammatory stimuli [97]. To elucidate which enzymatic function of PRDX6 might be responsible for the IL-6 secretion in senescent cells, we performed experiments with MJ33, an iPLA2 activity inhibitor [56]. These experiments indicated that the iPLA2 activity of PRDX6 might contribute to IL-6 secretion in IRIS, albeit the difference induced by the inhibitor was smaller than the effect of the knock-down.
Further experiments are necessary to provide more mechanistic details about the role of PRDX6 in IRIS. Based on the current knowledge we speculate that the different enzymatic activities of PRDX6 play distinctive functions in IRIS. Moreover, these functions are most likely specifically regulated and balanced as exemplified by the Cys47 hyperoxidation, which was lower than expected in a senescent model suffering from high ROS levels. The peroxidase activity might protect the cells from excessive oxidative damage and cell death, while the iPLA2 activity might play a role in senescence-associated inflammation.
5. Conclusion
In this work, we applied our previously developed SILAC-iodoTMT workflow to describe cysteine redox signaling events in IRIS. Our study suggests that the increase of ROS production in senescent cells does not cause any bulk cysteine oxidation. Instead, it results in specific changes in individual cysteine residues and augmentation of the expression of antioxidant proteins, suggesting mobilization of the antioxidant system rather than its exhaustion during the later stages in the IRIS. Further studies could shed light on the dynamics of cellular redoxome and proteome changes in different time points after the senescence-triggering treatment to fully elucidate the kinetics of cysteine oxidation in the establishment and maintenance of the senescent phenotype. Among the antioxidant proteins, PRDX6 is the key factor maintaining redox homeostasis and viability of senescent cells. Importantly, our study also provides a link between PRDX6 expression in IRIS and the development the pro-inflammatory secretory phenotype. Future studies should focus on linking the various enzymatic activities and subcellular localizations of PRDX6 to its functions in senescent cells. Moreover, PRDX6 activity could be exploited as a target of senolytic or senomodulatory drugs. A screening employing multiple cell lines and senescence-inducing mechanisms would further elucidate in what context such targeting would be efficient to eliminate senescent cells or mitigate the bystander effect of SASP. Altogether, we applied our novel redox detection method to determine the redox system response in IRIS development and to elucidate the complex role of PRDX6 in IRIS.
Author contributions
B.S., A.K., Z.L., M.V., and M.P. performed the experiments. B.S. performed the major data analysis. M.V., B.S., K.P., and I.F. acquired the LC-MS data. J.B. provided critical feedback. Z.H. and M.V. supervised the study. B.S., Z.H, and M.V. wrote the paper.
Data availability
The mass spectrometry proteomics data from this publication, MaxQuant and Spectronaut search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [110] with the data set identifiers PXD028772 and PXD028780 (http://proteomecentral.proteomexchange.org).
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This study was supported by the Grant Agency of the Czech Republic (GA20-15728S), and MH CZ DRO (UHHK, 00179906). B.S. was supported by the grant of the CAS (L200521953). We wish to thank Marketa Vancurova and Katerina Hruba from the Institute of Molecular Genetics of ASCR (Prague, Czech Republic) for their valued technical assistance and proof-reading. We acknowledge the Light Microscopy Core Facility, IMG CAS, Prague, Czech Republic, supported by MEYS (LM2018129, CZ.02.1.01/0.0/0.0/18_046/0016045) and RVO: 68378050-KAV-NPUI, for their support with the widefield imaging presented herein. The authors would like to acknowledge the Proteomics Service Laboratory at the Institute of Physiology (supported by RVO, ID 67985823) and Institute of Molecular Genetics (supported by RVO, ID 68378050) of the Czech Academy of Sciences for mass spectrometry proteomics analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102212.
Contributor Information
Marie Vajrychova, Email: marie.vajrychova@fnhk.cz.
Zdenek Hodny, Email: hodny@img.cas.cz.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Statistical analysis of the difference in cysteine oxidation status between senescent and proliferating cells based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the redox-proteomic analysis of senescent cells (cysteine oxidation relative quantification).
Results of the redox-proteomic analysis of senescent cells (relative protein quantification).
Enrichment analysis test results (1D enrichment analysis) of proteome changes in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Enrichment analysis test results (Fisher's exact test) of proteins up- or down-regulated in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the redox-proteomic analysis of senescent cells after PRDX6 knock-down (relative protein quantification).
Enrichment analysis test results (2D enrichment analysis) of proteome changes after PRDX6 knock-down in cellular senescence after based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Enrichment analysis test result (Fisher's exact test) of proteins up- or down-regulated after PRDX6 knock-down in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the secretome analysis of senescent cells after PRDX6 knock-down.
Multimedia component 10
References
- 1.Di Micco R., Krizhanovsky V., Baker D., d'Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021;22:75–95. doi: 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S., Schmitt C.A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019;21:94–101. doi: 10.1038/s41556-018-0249-2. [DOI] [PubMed] [Google Scholar]
- 3.Toussaint O., Medrano E.E., von Zglinicki T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp. Gerontol. 2000;35:927–945. doi: 10.1016/s0531-5565(00)00180-7. [DOI] [PubMed] [Google Scholar]
- 4.Toussaint O., Weemaels G., Debacq-Chainiaux F., Scharffetter-Kochanek K., Wlaschek M. Artefactual effects of oxygen on cell culture models of cellular senescence and stem cell biology. J. Cell. Physiol. 2011;226:315–321. doi: 10.1002/jcp.22416. [DOI] [PubMed] [Google Scholar]
- 5.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee A.C., Fenster B.E., Ito H., Takeda K., Bae N.S., Hirai T., Yu Z.X., Ferrans V.J., Howard B.H., Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J. Biol. Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 8.Kobashigawa S., Kashino G., Mori H., Watanabe M. Relief of delayed oxidative stress by ascorbic acid can suppress radiation-induced cellular senescence in mammalian fibroblast cells. Mech. Ageing Dev. 2015;146–148:65–71. doi: 10.1016/j.mad.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Atamna H., Paler-Martinez A., Ames B.N. N-t-butyl hydroxylamine, a hydrolysis product of alpha-phenyl-N-t-butyl nitrone, is more potent in delaying senescence in human lung fibroblasts. J. Biol. Chem. 2000;275:6741–6748. doi: 10.1074/jbc.275.10.6741. [DOI] [PubMed] [Google Scholar]
- 10.Ameziane-El-Hassani R., Talbot M., de Souza Dos Santos M.C., Al Ghuzlan A., Hartl D., Bidart J.M., De Deken X., Miot F., Diallo I., de Vathaire F., et al. NADPH oxidase DUOX1 promotes long-term persistence of oxidative stress after an exposure to irradiation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:5051–5056. doi: 10.1073/pnas.1420707112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubackova S., Krejcikova K., Bartek J., Hodny Z. IL1- and TGFbeta-Nox 4 signaling, oxidative stress and DNA damage response are shared features of replicative, oncogene-induced, and drug-induced paracrine 'bystander senescence. Aging (Albany NY) 2012;4:932–951. doi: 10.18632/aging.100520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyemi U., Lagente-Chevallier O., Boufraqech M., Prenois F., Courtin F., Caillou B., Talbot M., Dardalhon M., Al Ghuzlan A., Bidart J.M., et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2012;31:1117–1129. doi: 10.1038/onc.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weyemi U., Redon C.E., Aziz T., Choudhuri R., Maeda D., Parekh P.R., Bonner M.Y., Arbiser J.L., Bonner W.M. Inactivation of NADPH oxidases NOX4 and NOX5 protects human primary fibroblasts from ionizing radiation-induced DNA damage. Radiat. Res. 2015;183:262–270. doi: 10.1667/RR13799.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Correia-Melo C., Passos J.F. Mitochondria: are they causal players in cellular senescence? Biochim. Biophys. Acta Bioenerg. (BBA) 2015;1847:1373–1379. doi: 10.1016/j.bbabio.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Passos J.F., Saretzki G., Ahmed S., Nelson G., Richter T., Peters H., Wappler I., Birket M.J., Harold G., Schaeuble K., et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vizioli M.G., Liu T., Miller K.N., Robertson N.A., Gilroy K., Lagnado A.B., Perez-Garcia A., Kiourtis C., Dasgupta N., Lei X., et al. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 2020;34:428–445. doi: 10.1101/gad.331272.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhee S.G., Kang S.W., Chang T.S., Jeong W., Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 18.Soito L., Williamson C., Knutson S.T., Fetrow J.S., Poole L.B., Nelson K.J. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2011;39:D332–D337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton M.B., O'Connor K.M. Peroxiredoxins and the regulation of cell death. Mol. Cell. 2016;39:72–76. doi: 10.14348/molcells.2016.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee S.G., Kil I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 21.Wood Z.A., Poole L.B., Karplus P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 22.Chen J.-W., Dodia C., Feinstein S.I., Jain M.K., Fisher A.B. 1-Cys peroxiredoxin, a bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. J. Biol. Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 23.Fisher A.B. The phospholipase A(2) activity of peroxiredoxin 6. J. Lipid Res. 2018;59:1132–1147. doi: 10.1194/jlr.R082578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T.-S., Sundaresh C.S., Feinstein S.I., Dodia C., Skach W.R., Jain M.K., Nagase T., Seki N., Ishikawa K.-i., Nomura N., et al. Identification of a human cDNA clone for lysosomal type Ca2+-independent phospholipase A2 and properties of the expressed protein. J. Biol. Chem. 1997;272:2542–2550. doi: 10.1074/jbc.272.4.2542. [DOI] [PubMed] [Google Scholar]
- 25.Fisher A.B., Dodia C., Sorokina E.M., Li H., Zhou S., Raabe T., Feinstein S.I. A novel lysophosphatidylcholine acyl transferase activity is expressed by peroxiredoxin 6. JLR (J. Lipid Res.) 2016;57:587–596. doi: 10.1194/jlr.M064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arevalo J.A., Vazquez-Medina J.P. The role of peroxiredoxin 6 in cell signaling. Antioxidants. 2018;7 doi: 10.3390/antiox7120172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S.Y., Jo H.-Y., Kim M.H., Cha Y.-y., Choi S.W., Shim J.-H., Kim T.J., Lee K.-Y. H2O2-dependent hyperoxidation of peroxiredoxin 6 (Prdx6) plays a role in cellular toxicity via up-regulation of iPLA2 activity. J. Biol. Chem. 2008;283:33563–33568. doi: 10.1074/jbc.M806578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez-Medina J.P., Tao J.Q., Patel P., Bannitz-Fernandes R., Dodia C., Sorokina E.M., Feinstein S.I., Chatterjee S., Fisher A.B. Genetic inactivation of the phospholipase A2 activity of peroxiredoxin 6 in mice protects against LPS-induced acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;316:L656–L668. doi: 10.1152/ajplung.00344.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anwar S., Yanai T., Sakai H. Overexpression of peroxiredoxin 6 protects neoplastic cells against apoptosis in canine Haemangiosarcoma. J. Comp. Pathol. 2016;155:29–39. doi: 10.1016/j.jcpa.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 30.He Y., Xu W., Xiao Y., Pan L., Chen G., Tang Y., Zhou J., Wu J., Zhu W., Zhang S., et al. Overexpression of peroxiredoxin 6 (PRDX6) promotes the aggressive phenotypes of esophageal squamous cell carcinoma. J. Cancer. 2018;9:3939–3949. doi: 10.7150/jca.26041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pak J.H., Choi W.H., Lee H.M., Joo W.D., Kim J.H., Kim Y.T., Kim Y.M., Nam J.H. Peroxiredoxin 6 overexpression attenuates cisplatin-induced apoptosis in human ovarian cancer cells. Cancer Invest. 2011;29:21–28. doi: 10.3109/07357907.2010.535056. [DOI] [PubMed] [Google Scholar]
- 32.Grueso M.J.L., Valero R.M.T., Carmona H.B., Ruiz D.J.L., Peinado J., McDonagh B., Aguilar R.R., Ruiz J.A.B., Peña C.A.P. Peroxiredoxin 6 down-regulation induces metabolic remodeling and cell cycle arrest in HepG2 cells. Antioxidants. 2019;8:505. doi: 10.3390/antiox8110505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Grueso M.J., Lagal D.J., García-Jiménez Á F., Tarradas R.M., Carmona-Hidalgo B., Peinado J., Requejo-Aguilar R., Bárcena J.A., Padilla C.A. Knockout of PRDX6 induces mitochondrial dysfunction and cell cycle arrest at G2/M in HepG2 hepatocarcinoma cells. Redox biology. 2020;37:101737. doi: 10.1016/j.redox.2020.101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoops B., Argyropoulou V., Becker S., Ferté L., Kuznetsova O. Multiple roles of peroxiredoxins in inflammation. Mol. Cell. 2016;39:60–64. doi: 10.14348/molcells.2016.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benipal B., Feinstein S.I., Chatterjee S., Dodia C., Fisher A.B. Inhibition of the phospholipase A2 activity of peroxiredoxin 6 prevents lung damage with exposure to hyperoxia. Redox biology. 2015;4:321–327. doi: 10.1016/j.redox.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shanshan Y., Beibei J., Li T., Minna G., Shipeng L., Li P., Yong Z. Phospholipase A2 of peroxiredoxin 6 plays a critical role in cerebral ischemia/reperfusion inflammatory injury. Front. Cell. Neurosci. 2017;11:99. doi: 10.3389/fncel.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vénéreau E., Ceriotti C., Bianchi M.E. DAMPs from cell death to new life. Front. Immunol. 2015;6 doi: 10.3389/fimmu.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharapov M.G., Glushkova O.V., Parfenyuk S.B., Gudkov S.V., Lunin S.M., Novoselova E.G. The role of TLR4/NF-kappaB signaling in the radioprotective effects of exogenous Prdx6. Arch. Biochem. Biophys. 2021;702:108830. doi: 10.1016/j.abb.2021.108830. [DOI] [PubMed] [Google Scholar]
- 39.Ho J.N., Lee S.B., Lee S.S., Yoon S.H., Kang G.Y., Hwang S.G., Um H.D. Phospholipase A2 activity of peroxiredoxin 6 promotes invasion and metastasis of lung cancer cells. Mol. Cancer Therapeut. 2010;9:825–832. doi: 10.1158/1535-7163.MCT-09-0904. [DOI] [PubMed] [Google Scholar]
- 40.Rolfs F., Huber M., Gruber F., Böhm F., Pfister H.J., Bochkov V.N., Tschachler E., Dummer R., Hohl D., Schäfer M., et al. Dual role of the antioxidant enzyme peroxiredoxin 6 in skin carcinogenesis. Cancer Res. 2013;73:3460–3469. doi: 10.1158/0008-5472.CAN-12-4369. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt A., Schmitz W., Hufnagel A., Schartl M., Meierjohann S. Peroxiredoxin 6 triggers melanoma cell growth by increasing arachidonic acid-dependent lipid signalling. Biochem. J. 2015;471:267–279. doi: 10.1042/BJ20141204. [DOI] [PubMed] [Google Scholar]
- 42.Yun H.-M., Park K.-R., Lee H.P., Lee D.H., Jo M., Shin D.H., Yoon D.-Y., Han S.B., Hong J.T. PRDX6 promotes lung tumor progression via its GPx and iPLA2 activities. Free Radic. Biol. Med. 2014;69:367–376. doi: 10.1016/j.freeradbiomed.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Yun H.M., Park K.R., Park M.H., Kim D.H., Jo M.R., Kim J.Y., Kim E.C., Yoon D.Y., Han S.B., Hong J.T. PRDX6 promotes tumor development via the JAK2/STAT3 pathway in a urethane-induced lung tumor model. Free Radic. Biol. Med. 2015;80:136–144. doi: 10.1016/j.freeradbiomed.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 44.Zha X., Wu G., Zhang H., Yang Y., Zhang Y., Ma L. PRDX6 regulates the H2O2 and blue light-induced APRE-19 cell apoptosis via down-regulating and interacting with RARA. Anim. Cell Syst.(Seoul) 2019;23:241–245. doi: 10.1080/19768354.2019.1592021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zha X., Wu G., Zhao X., Zhou L., Zhang H., Li J., Ma L., Zhang Y. PRDX6 protects ARPE-19 cells from oxidative damage via PI3K/AKT signaling. Cell. Physiol. Biochem. : Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015;36:2217–2228. doi: 10.1159/000430186. [DOI] [PubMed] [Google Scholar]
- 46.Chhunchha B., Singh P., Stamer W.D., Singh D.P. Prdx6 retards senescence and restores trabecular meshwork cell health by regulating reactive oxygen species. Cell Death Dis. 2017;3:17060. doi: 10.1038/cddiscovery.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fatma N., Kubo E., Toris C.B., Stamer W.D., Camras C.B., Singh D.P. PRDX6 attenuates oxidative stress- and TGFbeta-induced abnormalities of human trabecular meshwork cells. Free Radic. Res. 2009;43:783–795. doi: 10.1080/10715760903062887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vajrychova M., Salovska B., Pimkova K., Fabrik I., Tambor V., Kondelova A., Bartek J., Hodny Z. Quantification of cellular protein and redox imbalance using SILAC-iodoTMT methodology. Redox biology. 2019;24:101227. doi: 10.1016/j.redox.2019.101227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 50.Debacq-Chainiaux F., Erusalimsky J.D., Campisi J., Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 51.Vancurova M., Hanzlikova H., Knoblochova L., Kosla J., Majera D., Mistrik M., Burdova K., Hodny Z., Bartek J. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair (Amst) 2019;78:114–127. doi: 10.1016/j.dnarep.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Victorelli S., Passos J.F. Reactive oxygen species detection in senescent cells. Methods Mol. Biol. 2019;1896:21–29. doi: 10.1007/978-1-4939-8931-7_3. [DOI] [PubMed] [Google Scholar]
- 54.Feoktistova M., Geserick P., Leverkus M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016 doi: 10.1101/pdb.prot087379. 2016(4):pdb.prot087379. [DOI] [PubMed] [Google Scholar]
- 55.Oleksak P., Psotka M., Vancurova M., Sapega O., Bieblova J., Reinis M., Rysanek D., Mikyskova R., Chalupova K., Malinak D., et al. Design, synthesis, and in vitro evaluation of BP-1-102 analogs with modified hydrophobic fragments for STAT3 inhibition. J. Enzym. Inhib. Med. Chem. 2021;36:410–424. doi: 10.1080/14756366.2020.1871336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fisher A.B., Dodia C., Chander A., Jain M. A competitive inhibitor of phospholipase A2 decreases surfactant phosphatidylcholine degradation by the rat lung. Biochem. J. 1992;288:407–411. doi: 10.1042/bj2880407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 58.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 59.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 60.Klammer M., Dybowski J.N., Hoffmann D., Schaab C. Identification of significant features by the global mean rank test. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox J., Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinf. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang da W., Sherman B.T., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Araki K., Kusano H., Sasaki N., Tanaka R., Hatta T., Fukui K., Natsume T. Redox sensitivities of global cellular cysteine residues under reductive and oxidative stress. J. Proteome Res. 2016;15:2548–2559. doi: 10.1021/acs.jproteome.6b00087. [DOI] [PubMed] [Google Scholar]
- 64.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smyth G.K. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Springer New York; New York, NY: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 66.Hebert A.S., Richards A.L., Bailey D.J., Ulbrich A., Coughlin E.E., Westphall M.S., Coon J.J. The one hour yeast proteome. Mol. Cell. Proteomics. 2014;13:339–347. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bekker-Jensen D.B., Martinez-Val A., Steigerwald S., Ruther P., Fort K.L., Arrey T.N., Harder A., Makarov A., Olsen J.V. A compact quadrupole-orbitrap mass spectrometer with FAIMS interface improves proteome coverage in short LC gradients. Mol. Cell. Proteomics. 2020;19:716–729. doi: 10.1074/mcp.TIR119.001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willforss J., Chawade A., Levander F. NormalyzerDE: online tool for improved normalization of omics expression data and high-sensitivity differential expression analysis. J. Proteome Res. 2019;18:732–740. doi: 10.1021/acs.jproteome.8b00523. [DOI] [PubMed] [Google Scholar]
- 69.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 70.Canovas B., Nebreda A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021;22:346–366. doi: 10.1038/s41580-020-00322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghezzi P., Chan P. Redox proteomics applied to the thiol secretome. Antioxidants Redox Signal. 2017;26:299–312. doi: 10.1089/ars.2016.6732. [DOI] [PubMed] [Google Scholar]
- 72.Go Y.-M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fisher A.B., Vasquez-Medina J.P., Dodia C., Sorokina E.M., Tao J.-Q., Feinstein S.I. Peroxiredoxin 6 phospholipid hydroperoxidase activity in the repair of peroxidized cell membranes. Redox biology. 2018;14:41–46. doi: 10.1016/j.redox.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., Holtz A., Shah S., Sharma V., Ferrucci L., et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coppé J.P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michishita E., Nakabayashi K., Suzuki T., Kaul S.C., Ogino H., Fujii M., Mitsui Y., Ayusawa D. 5-Bromodeoxyuridine induces senescence-like phenomena in mammalian cells regardless of cell type or species. J. Biochem. 1999;126:1052–1059. doi: 10.1093/oxfordjournals.jbchem.a022549. [DOI] [PubMed] [Google Scholar]
- 77.Davalli P., Mitic T., Caporali A., Lauriola A., D'Arca D. ROS, cell senescence, and novel molecular mechanisms in aging and age-related diseases. Oxidative Med. Cell. Longev. 2016:3565127. doi: 10.1155/2016/3565127. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 79.Anjo S.I., Melo M.N., Loureiro L.R., Sabala L., Castanheira P., Graos M., Manadas B. oxSWATH: an integrative method for a comprehensive redox-centered analysis combined with a generic differential proteomics screening. Redox biology. 2019:101130. doi: 10.1016/j.redox.2019.101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brandes N., Tienson H., Lindemann A., Vitvitsky V., Reichmann D., Banerjee R., Jakob U. Time line of redox events in aging postmitotic cells. eLife. 2013;2 doi: 10.7554/eLife.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray C.I., Van Eyk J.E. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ. Cardiovasc. Genet. 2012;5:591. doi: 10.1161/CIRCGENETICS.111.961425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parker J., Balmant K., Zhu F., Zhu N., Chen S. cysTMTRAQ-An integrative method for unbiased thiol-based redox proteomics. Mol. Cell. Proteomics. 2015;14:237–242. doi: 10.1074/mcp.O114.041772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shakir S., Vinh J., Chiappetta G. Quantitative analysis of the cysteine redoxome by iodoacetyl tandem mass tags. Anal. Bioanal. Chem. 2017;409:3821–3830. doi: 10.1007/s00216-017-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Topf U., Suppanz I., Samluk L., Wrobel L., Boser A., Sakowska P., Knapp B., Pietrzyk M.K., Chacinska A., Warscheid B. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018;9:324. doi: 10.1038/s41467-017-02694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Der Reest J., Lilla S., Zheng L., Zanivan S., Gottlieb E. Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao H., Jedrychowski M.P., Schweppe D.K., Huttlin E.L., Yu Q., Heppner D.E., Li J., Long J., Mills E.L., Szpyt J., et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell. 2020;180:968–983. doi: 10.1016/j.cell.2020.02.012. e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang J., Carroll K.S., Liebler D.C. The expanding landscape of the thiol redox proteome. Mol. Cell. Proteomics. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y., Luo Z., Wu X., Zhu J., Yu K., Jin Y., Zhang Z., Zhao S., Zhou L. Proteomic analyses of cysteine redox in high-fat-fed and fasted mouse livers: implications for liver metabolic homeostasis. J. Proteome Res. 2018;17:129–140. doi: 10.1021/acs.jproteome.7b00431. [DOI] [PubMed] [Google Scholar]
- 89.Menger K.E., James A.M., Cocheme H.M., Harbour M.E., Chouchani E.T., Ding S., Fearnley I.M., Partridge L., Murphy M.P. Fasting, but not aging, dramatically alters the redox status of cysteine residues on proteins in Drosophila melanogaster. Cell Rep. 2015;13:1285. doi: 10.1016/j.celrep.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcotte R., Lacelle C., Wang E. Senescent fibroblasts resist apoptosis by downregulating caspase-3. Mech. Ageing Dev. 2004;125:777–783. doi: 10.1016/j.mad.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 91.Sagiv A., Biran A., Yon M., Simon J., Lowe S.W., Krizhanovsky V. Granule exocytosis mediates immune surveillance of senescent cells. Oncogene. 2013;32:1971–1977. doi: 10.1038/onc.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang E. Senescent human fibroblasts resist programmed cell death, and failure to suppress bcl 2 is involved. Cancer Res. 1995;55:2284–2292. [PubMed] [Google Scholar]
- 93.Chang J., Wang Y., Shao L., Laberge R.M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W., et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yosef R., Pilpel N., Tokarsky-Amiel R., Biran A., Ovadya Y., Cohen S., Vadai E., Dassa L., Shahar E., Condiotti R., et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanders Y.Y., Liu H., Zhang X., Hecker L., Bernard K., Desai L., Liu G., Thannickal V.J. Histone modifications in senescence-associated resistance to apoptosis by oxidative stress. Redox biology. 2013;1:8–16. doi: 10.1016/j.redox.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Youn C.K., Song P.I., Kim M.H., Kim J.S., Hyun J.W., Choi S.J., Yoon S.P., Chung M.H., Chang I.Y., You H.J. Human 8-oxoguanine DNA glycosylase suppresses the oxidative stress induced apoptosis through a p53-mediated signaling pathway in human fibroblasts. Mol. Cancer Res. 2007;5:1083–1098. doi: 10.1158/1541-7786.MCR-06-0432. [DOI] [PubMed] [Google Scholar]
- 97.Kim S.Y., Chun E., Lee K.Y. Phospholipase A(2) of peroxiredoxin 6 has a critical role in tumor necrosis factor-induced apoptosis. Cell Death Differ. 2011;18:1573–1583. doi: 10.1038/cdd.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eismann T., Huber N., Shin T., Kuboki S., Galloway E., Wyder M., Edwards M.J., Greis K.D., Shertzer H.G., Fisher A.B., et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee D.H., Jung Y.Y., Park M.H., Jo M.R., Han S.B., Yoon D.Y., Roh Y.S., Hong J.T. Peroxiredoxin 6 confers protection against nonalcoholic fatty liver disease through maintaining mitochondrial function. Antioxidants Redox Signal. 2019;31:387–402. doi: 10.1089/ars.2018.7544. [DOI] [PubMed] [Google Scholar]
- 100.Ma S., Zhang X., Zheng L., Li Z., Zhao X., Lai W., Shen H., Lv J., Yang G., Wang Q., et al. Peroxiredoxin 6 is a crucial factor in the initial step of mitochondrial clearance and is upstream of the PINK1-parkin pathway. Antioxidants Redox Signal. 2015;24:486–501. doi: 10.1089/ars.2015.6336. [DOI] [PubMed] [Google Scholar]
- 101.Birch J., Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34:1565–1576. doi: 10.1101/gad.343129.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Faget D.V., Ren Q., Stewart S.A. Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer. 2019;19:439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 103.Davalos A.R., Kawahara M., Malhotra G.K., Schaum N., Huang J., Ved U., Beausejour C.M., Coppe J.-P., Rodier F., Campisi J. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 2013;201:613–629. doi: 10.1083/jcb.201206006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wiley C.D., Liu S., Limbad C., Zawadzka A.M., Beck J., Demaria M., Artwood R., Alimirah F., Lopez-Dominguez J.A., Kuehnemann C., et al. SILAC analysis reveals increased secretion of hemostasis-related factors by senescent cells. Cell Rep. 2019;28:3329–3337. doi: 10.1016/j.celrep.2019.08.049. e3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blokland K.E.C., Pouwels S.D., Schuliga M., Knight D.A., Burgess J.K. Regulation of cellular senescence by extracellular matrix during chronic fibrotic diseases. Clin. Sci. (London, England : 1979) 2020;134:2681–2706. doi: 10.1042/CS20190893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Valiente-Alandi I., Potter S.J., Salvador A.M., Schafer A.E., Schips T., Carrillo-Salinas F., Gibson A.M., Nieman M.L., Perkins C., Sargent M.A., et al. Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation. 2018;138:1236–1252. doi: 10.1161/CIRCULATIONAHA.118.034609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melhem H., Spalinger M.R., Cosin-Roger J., Atrott K., Lang S., Wojtal K.A., Vavricka S.R., Rogler G., Frey-Wagner I. Prdx6 deficiency ameliorates DSS colitis: relevance of compensatory antioxidant mechanisms. J. Crohn's & colitis. 2017;11:871–884. doi: 10.1093/ecco-jcc/jjx016. [DOI] [PubMed] [Google Scholar]
- 108.Min Y., Wi S.M., Shin D., Chun E., Lee K.-Y. Peroxiredoxin-6 negatively regulates bactericidal activity and NF-κB activity by interrupting TRAF6-ECSIT complex. Front. Cell. Infect. Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo J., Cao W., Chen C., Chen X. Peroxiredoxin 6 overexpression regulates adriamycin-induced myocardial injury, oxidative stress and immune response in rats. Ann. Transl. Med. 2020;8:1320. doi: 10.21037/atm-20-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis of the difference in cysteine oxidation status between senescent and proliferating cells based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the redox-proteomic analysis of senescent cells (cysteine oxidation relative quantification).
Results of the redox-proteomic analysis of senescent cells (relative protein quantification).
Enrichment analysis test results (1D enrichment analysis) of proteome changes in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Enrichment analysis test results (Fisher's exact test) of proteins up- or down-regulated in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the redox-proteomic analysis of senescent cells after PRDX6 knock-down (relative protein quantification).
Enrichment analysis test results (2D enrichment analysis) of proteome changes after PRDX6 knock-down in cellular senescence after based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Enrichment analysis test result (Fisher's exact test) of proteins up- or down-regulated after PRDX6 knock-down in cellular senescence based on their biological annotation (GOBP, GOCC, GOMF, Corum, KEGG).
Results of the secretome analysis of senescent cells after PRDX6 knock-down.
Multimedia component 10
Data Availability Statement
The mass spectrometry proteomics data from this publication, MaxQuant and Spectronaut search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [110] with the data set identifiers PXD028772 and PXD028780 (http://proteomecentral.proteomexchange.org).