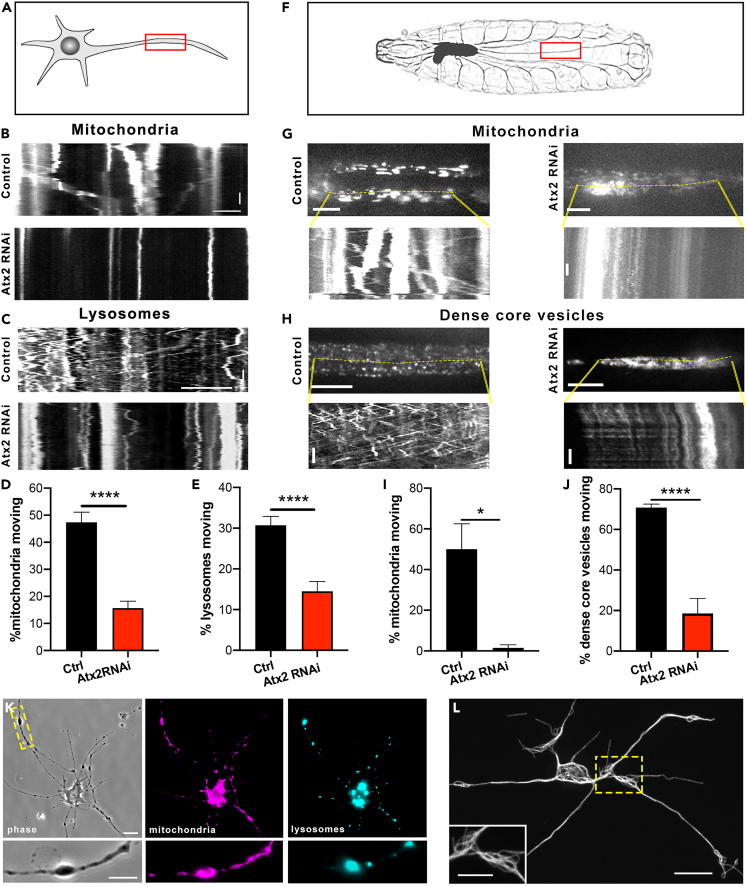
Figure 3.
Ataxin-2 depletion inhibits organelle transport
(A) Schematic showing region of imaging in neurons in culture.
(B) Example kymographs showing mitochondria motility in processes of control and elav > Atx2 RNAi neurons. Horizontal scale bar, 10μm. Vertical scale bar, 20s.
(C) Example kymographs showing lysosome motility in processes of control and Atx2 RNAi neurons. Horizontal scale bar, 10 μm. Vertical scale bar, 20s.
(D) Quantification of motile mitochondria in control and Atx2 knockdown neurons (Control = 47% ± 3.7, n = 25 cells, Atx2 RNAi = 15.7% ± 2.5, n = 28 cells, p < 0.0001).
(E) Quantification of motile lysosomes in control and Atx2 knockdown neurons (Control = 31% ± 2.1, n = 25 cells, Atx2 RNAi = 14.5% ± 2.4, n = 29 cells, p < 0.0001).
(F–H) (F) Schematic image of a third instar larva showing segmental nerves. Example kymographs showing mitochondria motility (G) and dense-core vesicle motility (H) in segmental nerves in vivo in control and elav > Atx2 RNAi larvae. Vertical scale bars, 10s, horizontal scale bars, 10 μm for dense-core vesicles, 20 μm for mitochondria.
(I) Quantification of motile mitochondria in control and Atx2 knockdown segmental nerves in vivo (Control = 50% ± 12.5, n = 3 larvae, Atx2 RNAi = 1.5% ± 1.5, n = 3 larvae, p = 0.015).
(J) Quantification of motile DCVs in control and Atx2 knockdown segmental nerves in vivo (Control = 66% ± 1.7, n = 9 larvae, Atx2 RNAi = 18.6% ± 7.4, n = 8 larvae, p < 0.0001).
(K) Example image showing mitochondria and lysosome distribution in an elav > Atx2 RNAi cultured neuron. Scale bar, 10 μm, inset 5 μm.
(L) Example super-resolution images of microtubules in varicosities in Atx2 RNAi neurons. Scale bar 10 μm, inset 5 μm. Data are presented as mean ± standard error. ∗p < 0.05, ∗∗∗∗p < 0.0001.