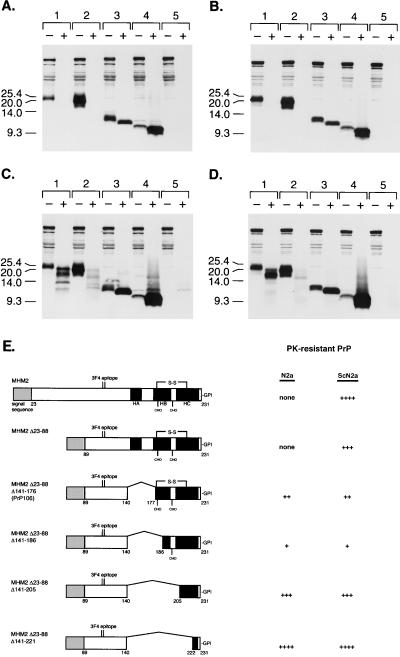
FIG. 2.
Expression of PrP deletion constructs in control and scrapie-infected neuroblastoma cells. Control (N2a) or scrapie-infected (ScN2a) cells were transfected transiently with the pSPOX expression vector carrying modified PrP genes as noted below. (A) Uninfected N2a cells digested with 20 μg of proteinase K per ml for 1 h at 37°C; (B) infected ScN2a cells digested with 20 μg of proteinase K per ml for 1 h at 37°C; (C) uninfected N2a cells digested with 7 μg of proteinase K per ml for 30 min at 37°C; (D) infected ScN2a cells digested with 7 μg of proteinase K per ml for 30 min at 37°C. Minus symbols denote undigested control sample, and plus symbols designate the pellet fraction of sample subjected to limited proteolysis as specified above. Samples that were digested with either 7 or 20 μg of proteinase K per ml correspond to a total protein/proteinase K ratio of 71:1 or 25:1, respectively. SDS-PAGE was performed on 16% Tricine gels (Novex). Western blotting was performed with 3F4 MAb as described in Materials and Methods. Paired sample lanes are numbered as follows: lane 1, MHM2(Δ23–88,Δ141–176), referred to as PrP106; lane 2, MHM2(Δ23–88,Δ141–186,C213A); lane 3, MHM2(Δ23–88,Δ141–205,C213A); lane 4, MHM2(Δ23–88,Δ141–221); lane 5, mock transfection. Units are apparent molecular sizes based on migration of protein standards in kilodaltons. (E) Schematic comparison of MHM2(Δ23–88) deletion mutants. Darkened areas correspond to α-helices determined in the nuclear magnetic resonance structure of PrP90 to -231 (10). None, no fragments detected with MAb 3F4 after digestion with 7 μg of proteinase K per ml for 30 min at 37°C; ++, fragments detected after digestion with 7 μg of proteinase K per ml for 30 min at 37°C but not after digestion with 20 μg of proteinase K per ml for 1 h at 37°C; ++++, intense 3F4 immunoreactive fragments seen even after digestion with 20 μg of proteinase K per ml for 1 h at 37°C. HA, α-helix A; HB, α-helix B; HC, α-helix C; CHO, carbohydrate; S-S, disulfide bond.