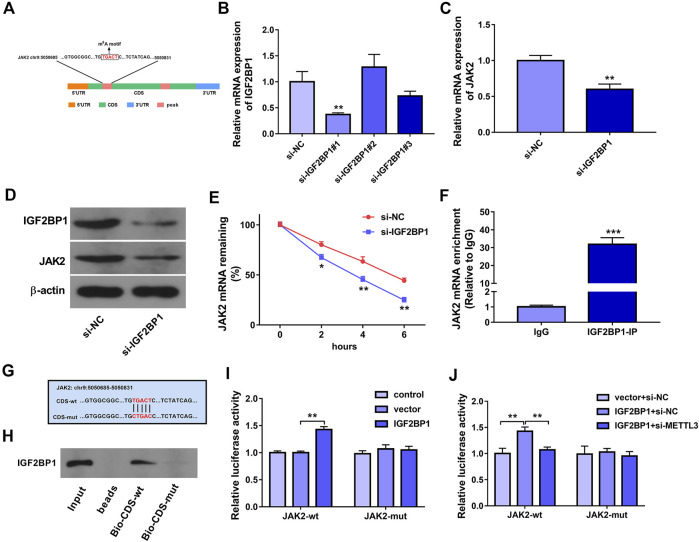
FIGURE 4.
IGF2BP1 positively regulated JAK2 expression and mRNA stability by directly binding to an m6A site within the CDS of JAK2 mRNA, and METTL3 knockdown impaired the interaction of IGF2BP1 and JAK2 mRNA in HUVECs. (A) The location of this m6A methylation motif in JAK2 mRNA. (B) HUVECs were transfected with si-NC, si-IGF2BP1#1, si-IGF2BP1#2, or si-IGF2BP1#3. Forty-eight hours later, IGF2BP1 mRNA level was measured through RT-qPCR assay. (C–E) HUVECs were transfected with si-NC or si-IGF2BP1#1 for 48 h (C) JAK2 mRNA level was measured through RT-qPCR assay. (D) Protein levels of IGF2BP1 and JAK2 were measured by western blot assay. (E) The stability of JAK2 mRNA was examined by ActD assay. (F) RIP assay was performed in HUVECs using the antibody against IgG or IGF2BP1. JAK2 mRNA level in IgG or IGF2BP1 immunoprecipitation complex was measured by RT-qPCR assay. (G) The wild or mutant m6A methylation site in JAK2 mRNA. (H) RNA pull-down assay was conducted in HUVECs using biotin-labeled JAK2 CDS-wt or CDS-mut probe. IGF2BP1 protein level pulled down by biotin-labeled probe was measured through western blot assay. (I, J) Luciferase activities were measured at 48 h after transfection. Three independent experiments were performed for all in vitro assays. *p < 0.05, **p < 0.01, ***p < 0.001.