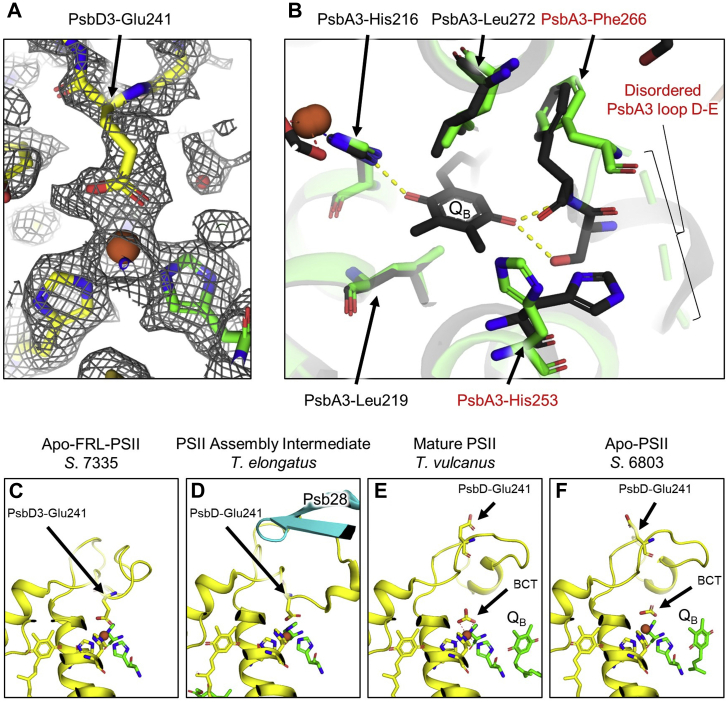
Figure 4.
Acceptor-side perturbations and comparison with other PSII structures.A, the apo-FRL-PSII model from Synechococcus 7335 is shown within the sharpened ESP map (9.5σ) focusing on PsbD3-Glu241 and its ligation to the NH-Fe. B, a structural superposition is shown of apo-FRL-PSII from Synechococcus 7335 (colored) and mature PSII from Thermosynechococcus vulcanus (black, PDB ID: 3WU2). Residues are labeled from the Synechococcus 7335 structure where red font denotes residues with significantly different positions compared with mature PSII. Note that the Ser side chain H-bonding to QB in mature PSII is PsbA-Ser264. The analogous residue in Synechococcus 7335 is PsbA3-Ser265, and it is not resolved in the ESP map (denoted by the dashed cartoon, labeled as “Disordered PsbA3 loop D–E”). Also note that all PsbA3 residues are numbered one greater in the Synechococcus 7335 PsbA3 sequence than they are in the sequence of T. vulcanus PsbA†. C–F, all panels show the cartoon representation of PsbD(3) and Psb28 when present. The stick representations of PsbD(3)-Glu241, QA, the NH-Fe-coordinating His residues, and QB and bicarbonate (labeled “BCT”) when present. C, the structure of apo-FRL-PSII from Synechococcus 7335 reported here. D, the structure of a PSII assembly intermediate from Thermosynechococcus elongatus (PDB ID: 7NHP). E, the structure of mature PSII from T. vulcanus (PDB ID: 3WU2). F, the structure of apo-PSII from Synechocystis sp. PCC 6803 (PDB ID: 6WJ6). ESP, electrostatic potential; FRL, far-red light; NH-Fe, non-heme Fe(II); PDB, Protein Data Bank; PSII, photosystem II.