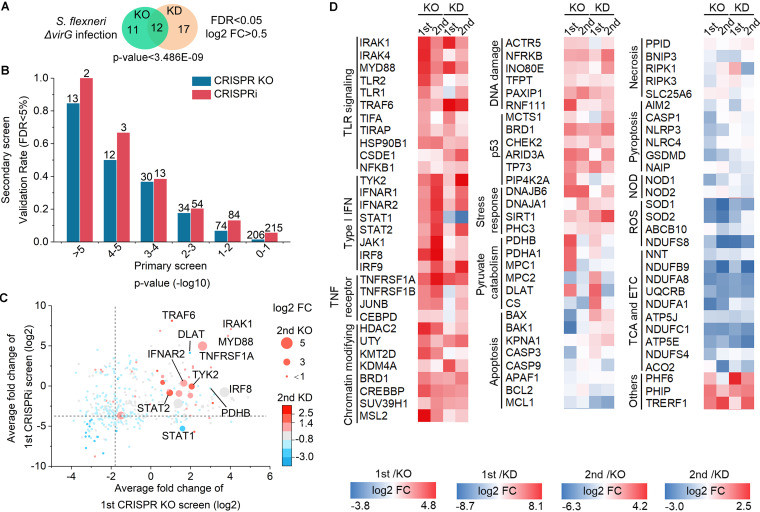
FIG 3.
Secondary CRISPR knockout and CRISPRi screens identify host genetic hits in S. flexneri infection. (A) Enriched genes were filtered with a cutoff FDR of <0.05 and a log2 fold change of >0.5 in S. flexneri ΔvirG infection. The degree of significance of the overlap is given. (B) Validation rates of genetic hits in the secondary screen grouped by their P values in the genome-wide screens in S. flexneri ΔvirG infection. The number of genes per category is indicated. (C) Genetic hits from both primary genome-wide and secondary screens were ranked by their differential sgRNA abundances between S. flexneri-infected and uninfected populations (log2 fold change). (D) Heatmap of screen hits clustered in different biological pathways in S. flexneri ΔvirG infection. ETC, electron transport chain; ROS, reactive oxygen species; NOD, nucleotide binding oligomerization domain.