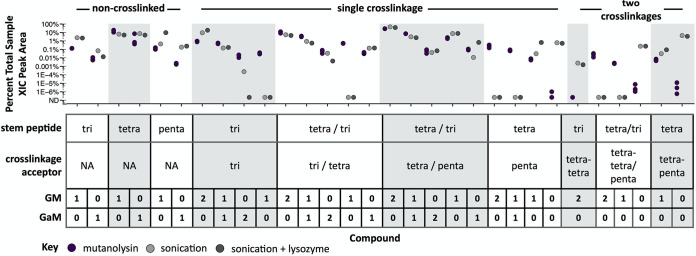
FIG 2.
Composition of Buchnera-derived cell wall fragments purified from A. pisum. Aphid homogenate was successively filtered to components of ≤10 kDa and subjected to LC-MS/MS analysis. Muropeptide compounds were identified from MS/MS spectra and extracted-ion chromatogram (XIC) peak areas determined using Byonic and Byologic softwares, respectively (Protein Metrics). Areas were baseline subtracted and normalized by sample, such that the data shown is the percentage of total PGN represented by each compound. Untreated mutanolysin-derived E. coli muropeptides are shown for comparison (purple). Two treatments were used without replicates: aphid homogenate was sonicated to lyse Buchnera cells (light gray) or additionally treated with lysozyme to digest Buchnera cell walls (dark gray). The table describes the compound structure: PGN compounds vary by stem peptide sequence and glycan (GM = GlcNAc-MurNAc, GaM = GlcNAc-anhydro-MurNAc). For distinct compounds that are equivalent in mass (differing either in stem peptide sequence or cross-linkage type), we were unable to quantify each compound abundance independently, because the two compounds could not be chromatographically resolved—such structural isomers were integrated together, and their sequences are reported with variable residues shown separated by backslashes within parentheses, such that the same relative position within parentheses refers to the sequence of one isomer.