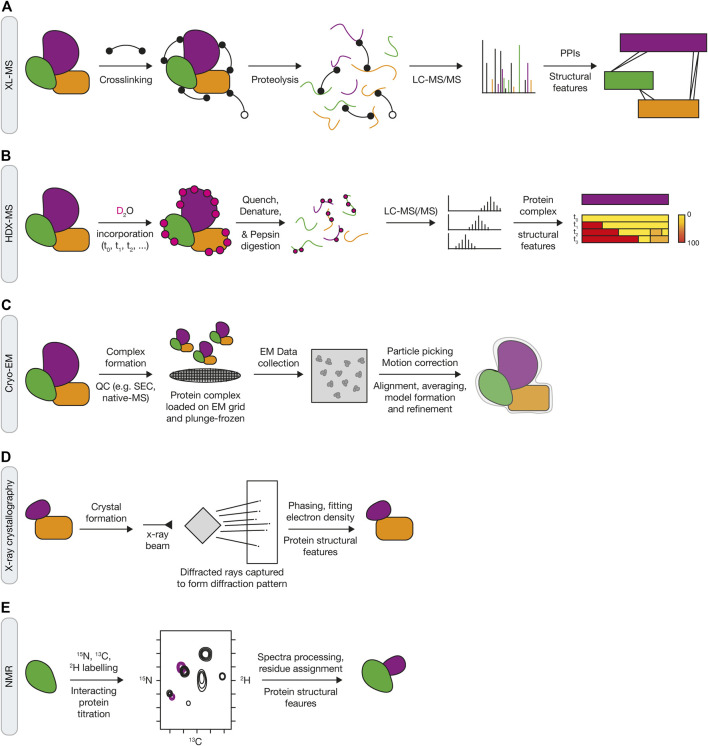
FIGURE 6.
Integrated structural approaches to investigate mechanisms of ubiquitin signaling in the DDR. (A) Crosslinking with mass spectrometry (XL-MS) requires the use of chemical crosslinkers to react with amino acid side chains, such as BS3 NHS-ester chemistry for primary amines on lysine residues or N-termini of proteins. Subsequent proteolysis with Trypsin or LysC, tandem mass spectrometry and data analysis using specialised software enables identification of crosslinked peptide species. (B) Hydrogen-deuterium mass spectrometry (HDX-MS) relies on the incorporation of 2H into the protein by incubation in deuterated water. The experiment uses a time course of 2H-incorporation followed by a rapid quenching and denaturation step at pH 2.5 before pepsin digestion and mass spectrometry analysis. Specialised data analysis pipelines can assess differences in 2H-incorporation for a protein across experimental conditions. (C) Single particle cryo-electron microscopy (cryo-EM) has recently evolved as a technique to get high-resolution structural data of larger multi-protein assemblies. Protein samples go through several stages of quality control (QC) via biochemical and biophysical techniques (e.g., SEC and native mass spectrometry) before loading onto carbon-coated EM grids, plunge-freezing in liquid ethane and data collection using high-power electron microscopes. (D) X-ray crystallography and (E) nuclear magnetic resonance (NMR) are established techniques to gain atomic-level resolution of protein structures that relies on the formation of crystals and isotopically labelled proteins, respectively.