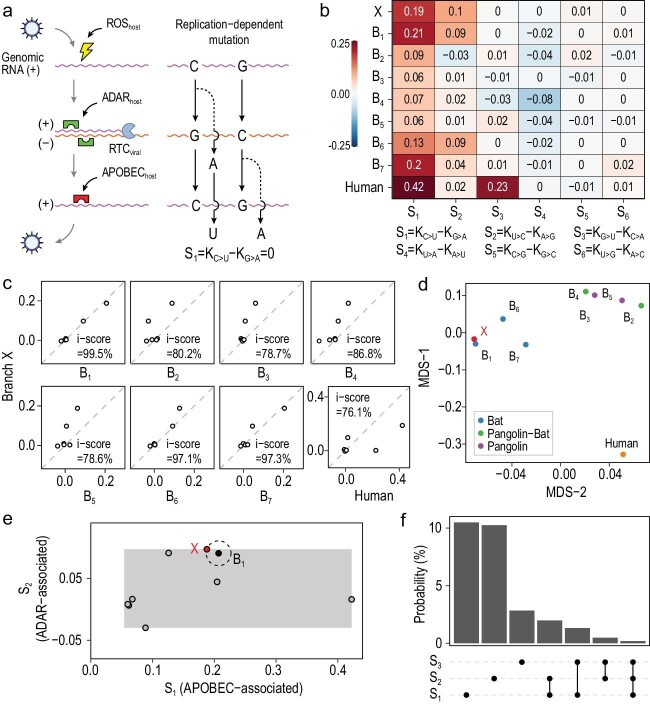
Figure 2.
Host signatures inferred from the viral mutation spectrum. (a) A diagram showing the major sources of viral mutations, which include the replication errors (by the viral replication-transcription complex, RTC) and the lesions caused by host factors. Because replication processes are the same, despite being in the opposite order, for nucleotides G and C (or A and T), replication errors would result in equal rates of complementary mutations such as C > A and G > T. However, host factors would distort the equal-rate pattern of complementary mutation pairs. The positive-sense RNA is often in a single-stranded form, sensitive to ROS and the APOBEC family, while the negative-sense RNA tends to be in a double-stranded form, thus more affected by the ADAR family. (b) The rate difference of each complementary mutation pair serves as a signature of host factors. There are thus six host signatures, each corresponding to a complementary mutation pair, inferred from the viral mutation spectrum. Among the three major host signatures, S1 is likely associated with the APOBEC family, S2 the ADAR family and S3 the ROS. (c) The similarity of host signatures between branch X and each of the other eight branches. Branch X is highly similar to B1, B6 and B7, the three branches of bat coronavirus. (d) A multidimensional scaling (MDS) plot of the host signatures reveals that branch X and B1 have nearly the same positions. (e) Estimation of the likelihood that an arbitrary laboratory condition happens to match the host signatures of B1 (the branch of RaTG13). The grey rectangular area is defined by the empirical ranges of S1 (APOBEC-associated) and S2 (ADAR-associated) that are based on the data of panel (b). The probability of approaching B1 as closely as X is the area of the circle divided by the whole rectangular area, which is ∼2.0%. The positions of the other seven branches are also shown in the rectangular area. (f) The probability that an arbitrary condition approaches B1 as closely as X is given, by considering the different combinations of S1, S2 and S3, respectively.