Fig. 4.
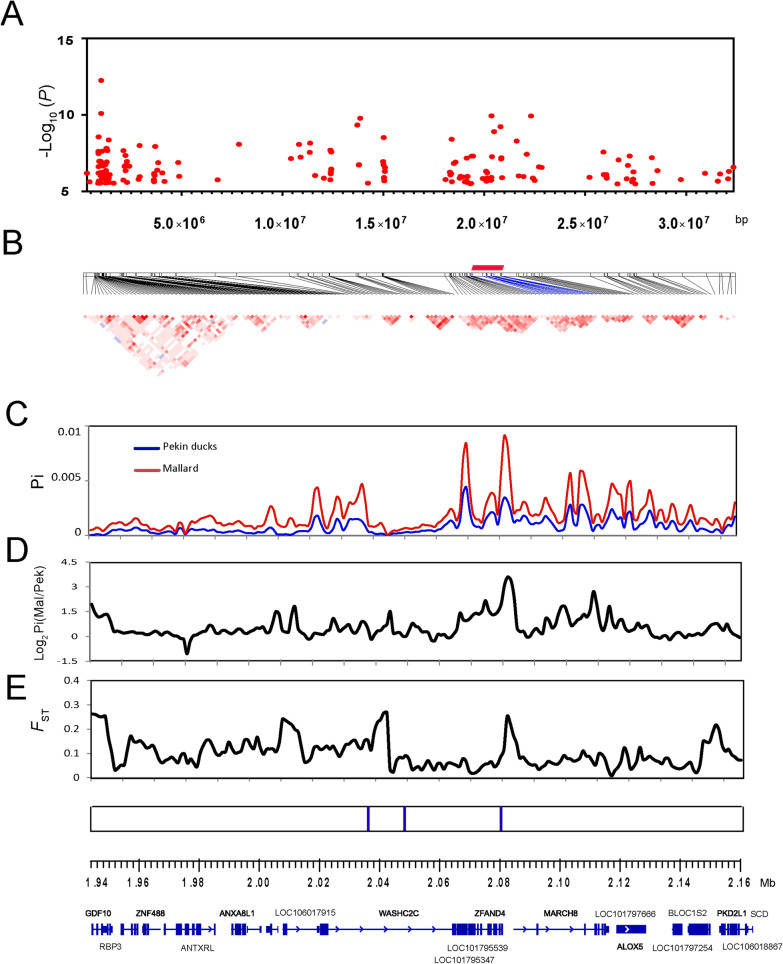
Analyses of the signatures of selection of the candidate region on chromosome 6 that affects FCR. a GWAS peaks located on chromosome 6. b Linkage disequilibrium (LD) of the significant SNPs. The red rectangle represents the region in LD with the leader SNP from the GWAS (Chr6:20,356,803, P = 1.14 × 10–10). c Average nucleotide diversities (Pi-value) of SNPs located in the sliding windows of the candidate region. d log2 Pi (mallard/Pekin duck) of the sliding windows. e Population differentiation analysis. f Each vertical blue line represents an SNP that reached the significance threshold of − log10 P = 8.16. The annotated genes are located in the candidate region according to the newly updated duck reference genome (IASCAAS_Peking Duck_PBH1.5, GCF_003850225.1)