FIGURE 1.
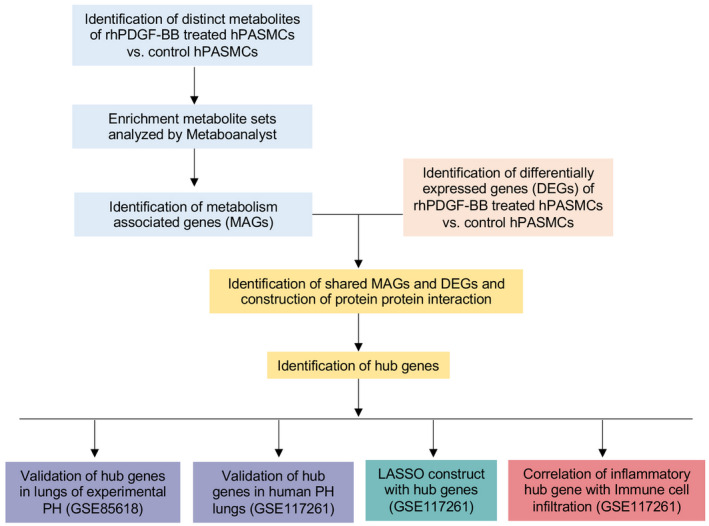
Main analysis flowchart. Identification of distinguishing metabolites of recombinant human recombinant PDGF‐BB (rhPDGF‐BB) treated human pulmonary arterial smooth muscle cells (hPASMCs) versus vehicle treated PASMCs. Enrichment of metabolite sets were analyzed by Metaboanalyst. Top 10 enriched pathways were identified and selected for further identification of metabolism associated genes (MAGs). Next, the shared MAGs and differentially expressed genes (DEGs) in rhPDGF‐BB treated hPASMCs versus control cells were identified. Protein protein interaction were constructed. Hub gene were identified by cytoHubba or MCODE add‐ins in Cytoscape. Common hub genes were validated in lungs of PH rats after hypoxia exposure from dataset GSE85618 and in lungs of PH patients from dataset GSE117261. LASSO regression model was constructed based on hub genes for PH prediction. The correlation of immune cell infiltration in lung tissues of dataset GSE117261 with inflammatory hub gene was also investigated
