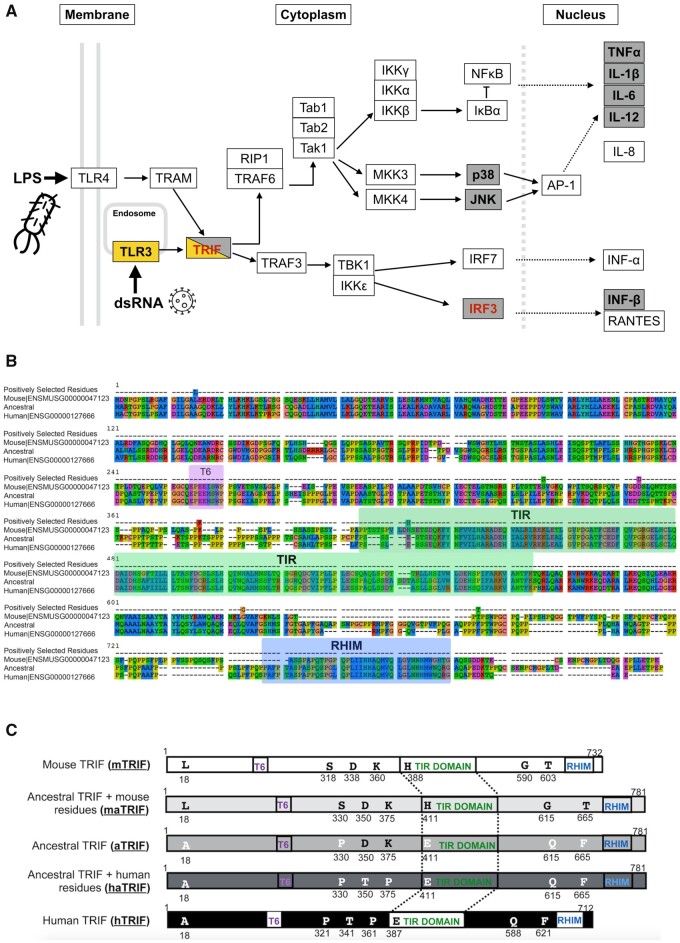
Fig. 1.
Signatures of adaptive evolution identified in TRIF and TRIF-dependent signaling pathways. (A) Schematic adapted from the Kyoto Encyclopedia of Genes and Genomes (KEGG) illustrating TRIF-dependent pathways in toll-like signaling in innate immunity. Selective pressure variation analysis was performed for both lineage-specific (model A) and site-specific (model 8) selection. Genes boxed in gray represent those that are predicted to have signatures of positive selection; gene names in bolded black contain residues that are predicted to be under site-specific selection, and genes in red are genes under selection in the mouse lineage only. Highlighted in yellow are TLR3 and TRIF predicted to be under selection in mouse by our earlier analysis (Webb et al. 2015). (B) A portion of the PRANK amino acid alignment of TRIF homologues showing the human, mouse, and ancestral TRIF protein sequences. The positions of the positively selected sites are indicated, as well as the position (relative to human—O'Neill and Bowie 2007) of the three known functional TRIF domains: Traf6 binding motif (T6), Toll/interleukin-1 receptor (TIR) domain, and the receptor-interacting protein (RIP) homotypic interaction motif (RHIM). (C) Schematic illustrating the different protein sequences of TRIF used in this study. Residues highlighted in the mouse protein (mTRIF) are those predicted to be under positive selection. Their position relative to the three functional domains is indicated. The position and identity of the homologous residues in the hTRIF, the reconstructed ancestral TRIF (aTRIF), the humanized ancestral TRIF protein (haTRIF), and the murinized ancestral TRIF (maTRIF) are also shown.