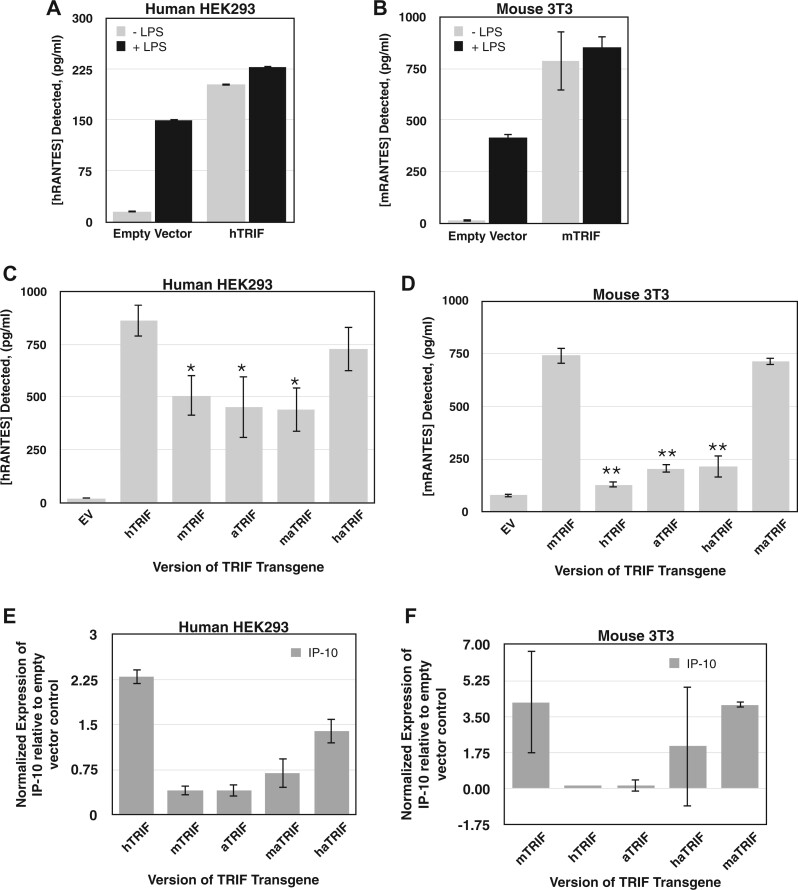
Fig. 2.
Amino acid residues under positive selection in TRIF, modulate species-specific function. Functional analysis of TRIF gene constructs. TRIF-induced RANTES production for human HEK 293 cells (A + C) and mouse 3T3 cells (B + D). Cell lines were transiently transfected with plasmids constitutively expressing each of the versions of the TRIF transgenes as indicated, or an empty vector (EV) control. In (A) and (B), cells were stimulated with LPS 12 h post-infection. At 24 h post transection, supernatants were recovered and assayed for the presence of the RANTES cytokine by ELISA. Each graph represents data from three biological replicates assayed in triplicate from a single day’s experiment. Similar trends were observed on different days. Error bars denote SD. *P < 0.05 compared with hRANTES value in HEK cells. **P < 0.01 compared with mRANTES value in 3T3 cells. Quantitative RT-PCR analysis as outlined in figure indicating the expression of IP-10 in human HEK 293 TLR4 cells (E) and in murine 3T3 cells (F) expressing the indicated TRIF constructs. Baseline of 0 is set to the expression of IP-10 in cells transfected with the empty vector control.