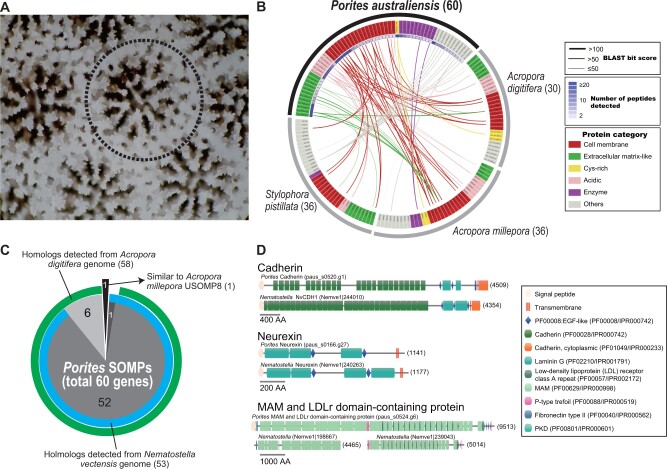
Fig. 2.
Skeletal proteome analysis of Porites australiensis. (A) High-magnification photo of a P. australiensis skeleton. The dotted line indicates a polyp. (B) A circos plot showing sequence similarities of PoritesSOMP genes against known SOMPs of other corals, Acropora digitifera, A. millepora, and Stylophora pistillata. Each box along the arc indicates a SOMP gene, and gene names are colored based on protein categories (cell membrane, extracellular matrix-like, cysteine-rich, acidic, enzymes, and others), as shown at the right. SOMPs connected by lines show significant sequence similarity (BLASTP, E-value ≤e−5), and thicknesses of lines reflect BLAST bit scores. Numbers of peptides detected by LC–MS/MS are shown by heatmap inside Porites gene names. (C) A pie chart showing numbers of putative homologs of PoritesSOMP genes (total 60) detected in Acropora digitifera (green) and sea anemone N. vectensis (blue) genomes. One gene (paus_s0004.g93) that was not detected in the two genomes showed similarity to A. milleporaUSOMP8 (Ramos-Silva et al. 2013). (D) Comparison of functional protein domain architectures of Porites cell membrane SOMPs, cadherin, neurexin, and MAM and LDLr dcps (upper) with their putative orthologs in Nematostella (lower). The amino acid length of each gene is shown in brackets.