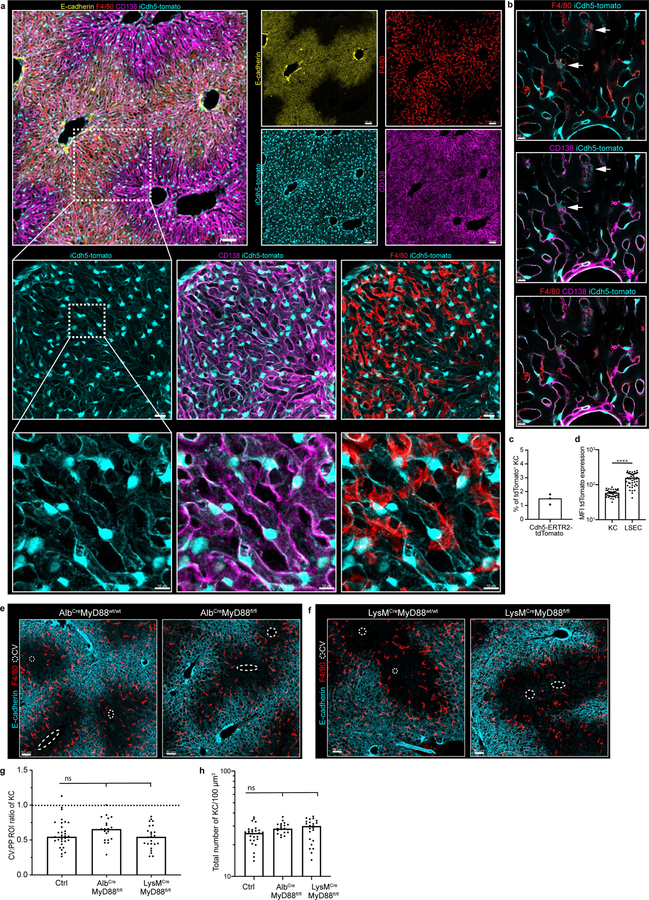
Extended Figure 4. iCdh5-Cre. Alb-Cre and LysM-Cre livers reporters: loss of MyD88 signalling in hepatocytes and KCs does not alter KC localization.
(a) Representative IF image showing iCdh5-Cre expression when crossed to a Rosa26-tdTomato reporter. (b) Zoomed in image: iCdh5-Cre-tdTomato is predominantly found on CD138+ LSECs, but can also be seen on a small number of KCs. (c) Percentage of total tdTomato+ KCs in iCdh5-Cre-tdTomato animals, each dot represents a biological replicate, n=3 mice. (d) MFI of tdTomato on tdTomato+ F4/80+ KCs and tdTomato+ LSECs from iCdh5-Cre-tdTomato (each dot represents a cell), n = 3 mice/condition, Two-tailed Mann-Whitney test, **** p<0.0001; Median shown. (e-f) Representative IF image showing F4/80+ KC distribution of (e) Alb-MyD88−/− and (f) LysM-MyD88−/− animals with littermate controls, CV highlighted by dashed circles in IFs. (g) Ratio of total KC numbers in CV to PP ROIs, (each dot represents a lobule), n = 4 mice/condition, Kruskal-Wallis test with Dunn’s multiple comparison test. For WT vs LysMCre-MyD88flfl-: ns p-value>0.9999, for WT vs AlbCre-MyD88fl/fl: ns p-value=0.2082; Median shown. (h) total KC numbers per volume; each dot represents a lobule. n = 4 mice/condition, Kruskal-Wallis test with Dunn’s multiple comparison test. For WT vs LysMCre-MyD88flfl-: ns p-value=0.0781, for WT vs AlbCre-MyD88fl/fl: ns p-value=0.0624; Median shown. Channels shown labelled in figures, CV highlighted by dashed circles in IFs.