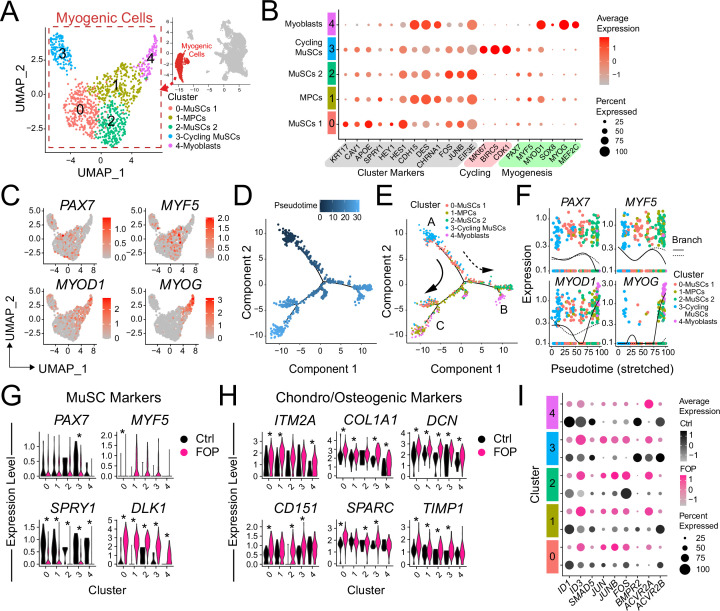
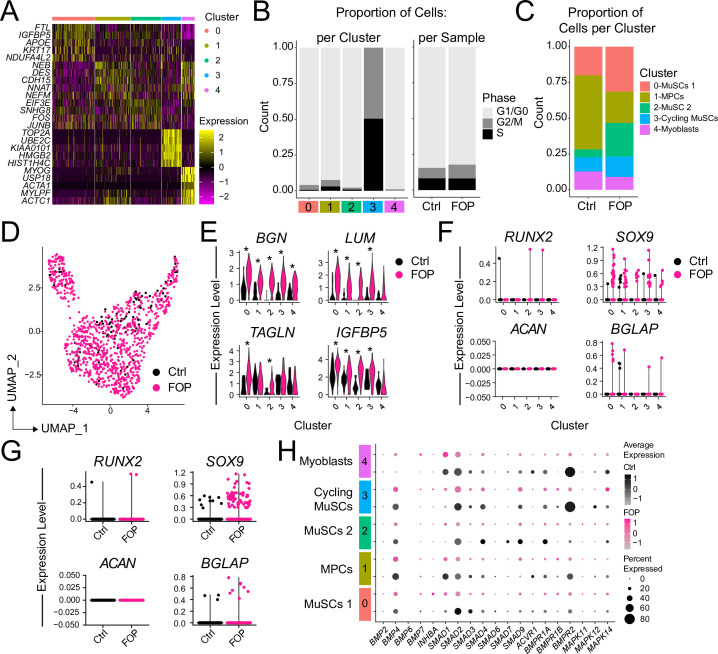
Figure 5. Transcriptional profile of iMPCs.
(A) Identified myogenic clusters (in red) were sub-clustered from the other clusters (neurogenic and mesenchymal) and re-analyzed. UMAP of the new myogenic subclusters was generated. (B) Dot plot displaying expression of cluster defining genes. (C) Feature expression plots of myogenic markers. (D) Myogenic cells were ordered as a function of pseudotime using the Monocle package. (E) Pseudotime trajectory plot of myogenic clusters.Arrows represents the direction and major branches of the pseudotime. Branches A-C are marked with letters along the trajectories. (F) Gene plots displaying the expression of specific myogenic genes as a function of pseudotime. (G–H) Violin plots of myogenic (G) and chondro/osteogenic genes (H) that were significantly differentially expressed. Violin plot width depicts the larger probability density of cells expressing each particular gene at the indicated expression level. *, significantly different following differential expression testing using the Wilcoxon rank sum test per cluster. (I) SMAD and P38MAPK pathway markers significantly differentially expressed between control and FOP myogenic cells. p values for (G–H) are in Figure 5—source data 1.