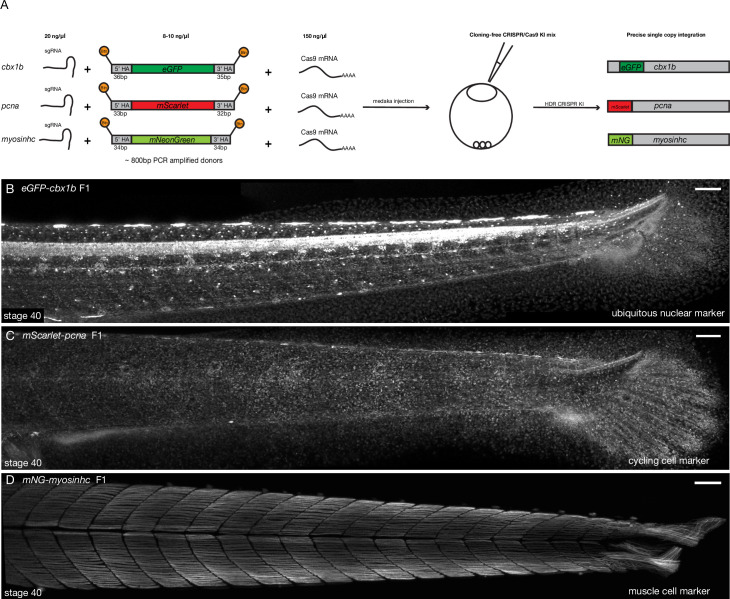
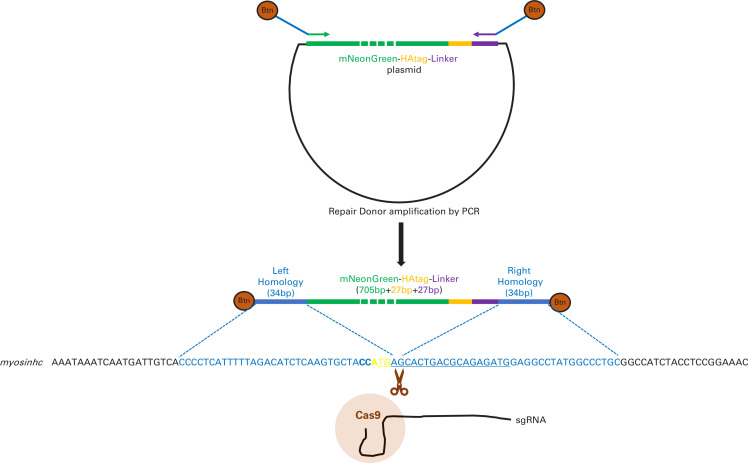
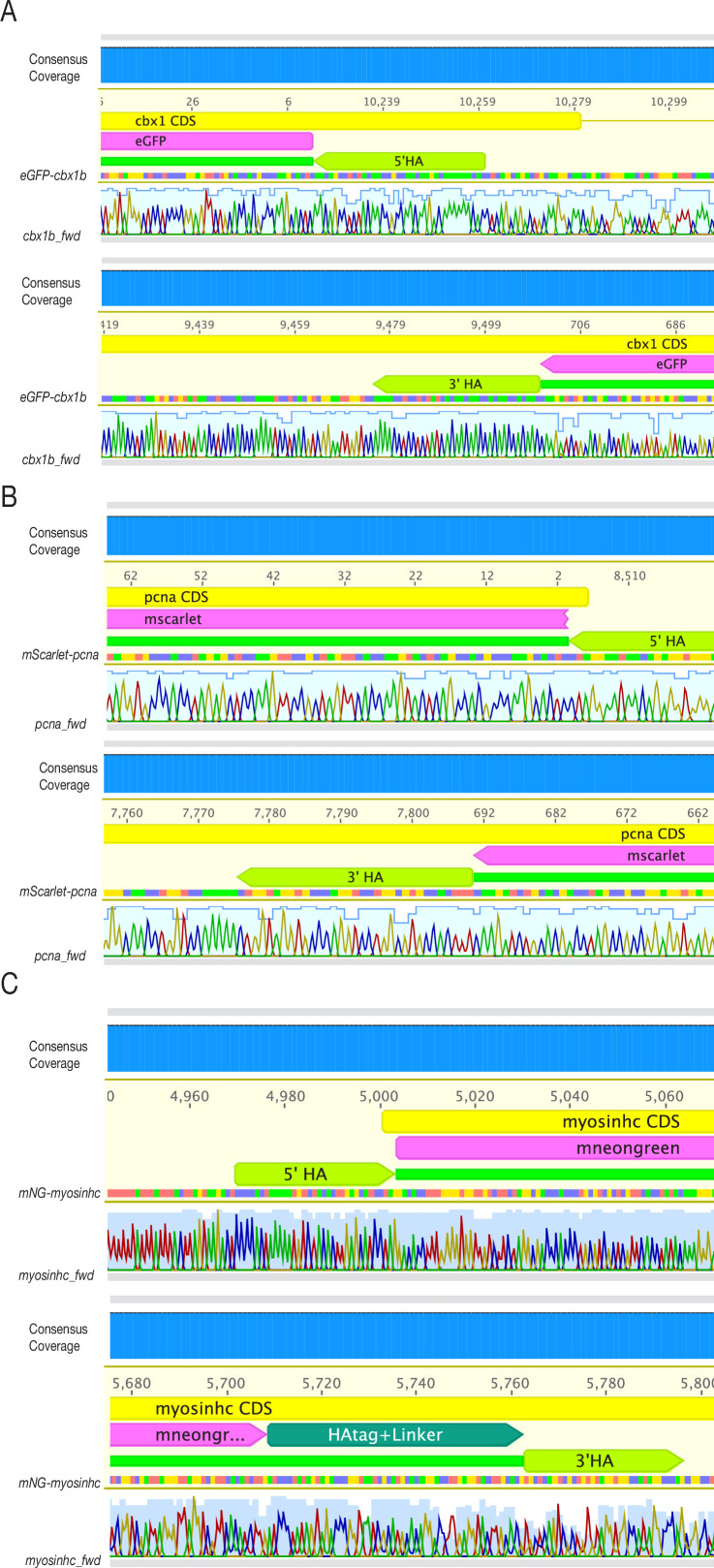
Figure 1. Cloning-free single-copy CRISPR/Cas9-mediated knock-in (KI) lines in medaka.
(A) Schematic diagram of cloning-free CRISPR knock-in strategy. The injection mix consists of three components, a single-guide RNA (sgRNA) targeting the gene of interest, Cas9-mSA mRNA, and the PCR-amplified donor fragment containing short homology arms on both ends (30–40 bp) and the fluorescent protein of interest with no ATG and no stop codon. Note that the 5′ ends of the PCR donor fragment are biotinylated (Btn). The mix is injected in one-cell staged medaka embryos and the injected fishes are screened for potential in-frame integrations mediated by homology-directed repair (HDR). (B) eGFP-cbx1b F1 CRISPR KI line stage 40 medaka embryos. eGFP-Cbx1b labels all nuclei and is thus an ubiquitous nuclear marker. n > 10 embryos. Scale bar = 100 µm (C) mScarlet-pcna F1 CRISPR KI line stage 40 medaka embryos. mScarlet-Pcna labels exclusively cycling cells. n > 10 embryos. Scale bar = 100 µm. (D) mNG-myosinhc F1 CRISPR KI line stage 40 medaka embryos. mNG-Myosinhc labels exclusively muscle cells located in the myotome tissue of medaka embryos. n > 10 embryos. Scale bar = 100 µm.