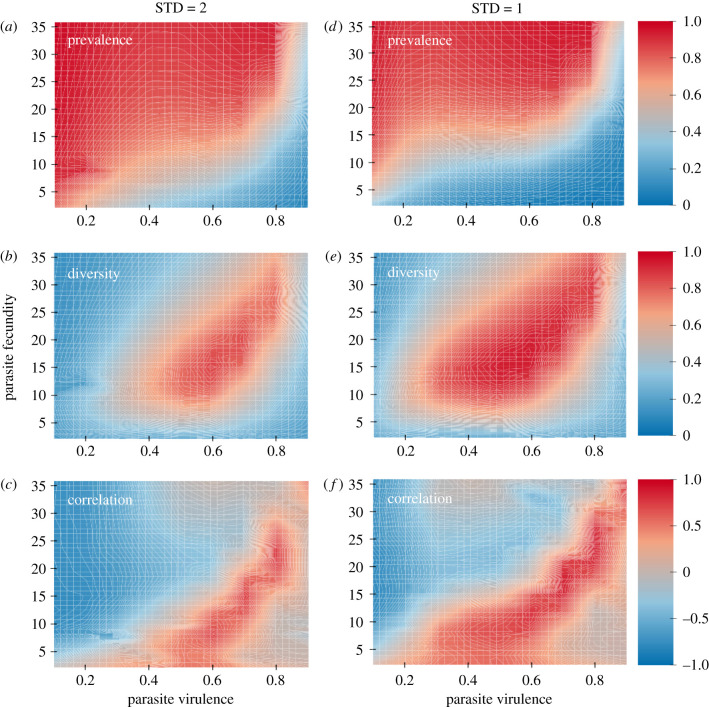
Figure 1.
Simulation results. The left-hand column (a–c) gives simulation results for which the standard deviation in host fecundity was 2.0, while the right-hand column (d–f) gives results for a standard deviation in host fecundity of 1.0. The higher standard deviation (a–c) gives stronger fecundity selection. The top row (a,d) gives the results for mean infection prevalence. The middle row (b,e) gives the results for mean diversity, while the lowest row (c,f) gives the correlation between diversity and prevalence of infection. The side bars show the values of the different colours for prevalence (a,d), diversity (b,e) and the correlation between diversity and prevalence (c,f). For graphs c and f, correlations having an absolute value of greater than 0.45 have p-values less than 0.05. Note that, as expected, high levels of diversity are maintained across more of the parameter space when fecundity selection is weaker. Note also that high correlations are shifted to the south and west relative to regions of high host diversity. As such, high levels of host diversity can be associated with low (or even negative) correlations between diversity and prevalence. Note that the simulation results assumed clonal reproduction, but they could also be applicable to highly inbred populations.