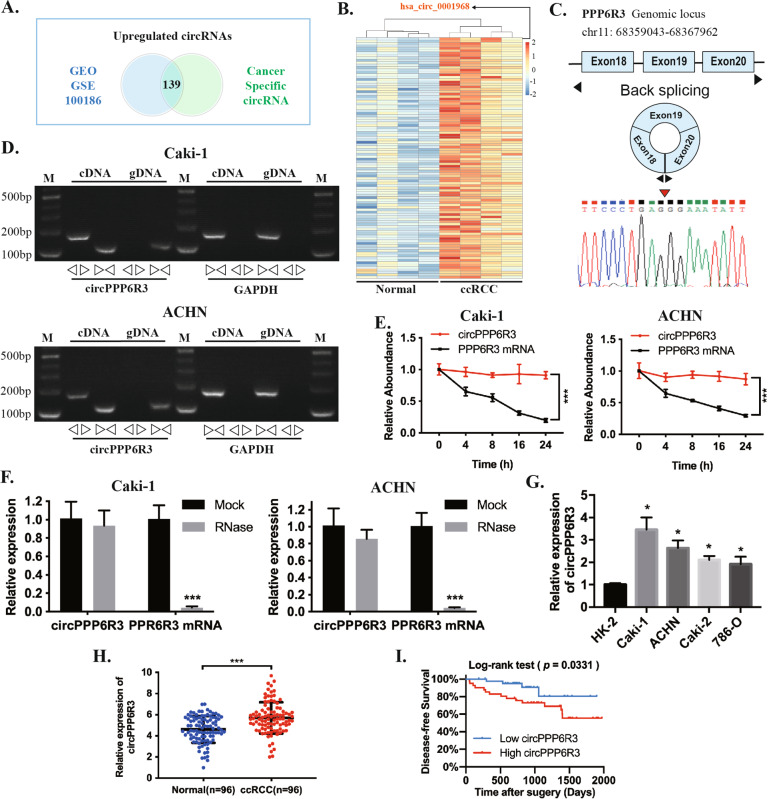
Fig. 1. circRNA expression profiles in ccRCC and characteristics of circPPP6R3.
A The upregulated circRNAs were screened by two online databases, GSE100186 comprised of four paired ccRCC tissues and normal tissues in the GEO database as well as the Cancer-Specific CircRNA database. B The cluster heatmap was conducted to present the top 100 upregulated circRNAs of this intersection in the GSE100186 dataset. C circPPP6R3 was produced by back-spliced of the exon 18, 19, and 20 of the PPP6R3 gene identified by sanger sequencing. D The PCR products of circPPP6R3 and its linear gene PPP6R3 mRNA from the cDNA and gDNA of ccRCC cells were tested by Northern Blotting, GAPDH was used as a control. E Actinomycin D was applied to detect the stability of circPPP6R3 and PPP6R3 mRNA. F The expression of circPPP6R3 and PPP6R3 mRNA were validated by qRT-PCR under the digestion of RNase R in ccRCC cells. G The expression of circPPP6R3 was determined by qRT-PCR in ccRCC cell lines and the renal tubular epithelial cell line, HK-2. H qRT-PCR analysis of circPPP6R3 in 96 paired of ccRCC tissues and adjacent normal tissues. I The Kaplan–Meier analysis indicated that the ccRCC patients with high expression of circPPP6R3 had unfavorable disease-free survival rates compared with low circPPP6R3 expression.