Fig. 2. The microbial diversity in the samples from the sediment cores.
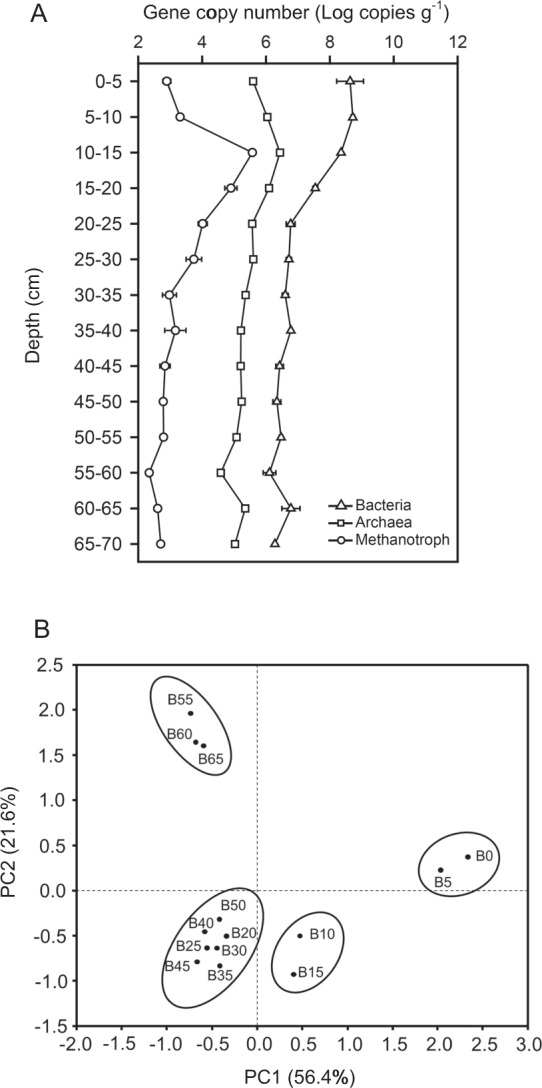
The abundance of bacteria, archaea, and methanotrophs estimated using quantitative real-time PCR (qPCR) targeting bacterial and archaeal 16S rRNA genes and pmoA based on the dry weight of sediment (n = 3) (A), and principal component analysis of bacterial terminal restriction fragment length polymorphism (T-RFLP) profiles of 16S rRNA gene amplicons digested with HhaI for DNA from the original sediment of 0–70 cm at 5 cm intervals (B), which were collected in ice-free July 2009. The number embedded within the sample name (i.e., B0, B5, B10, etc.) from 0 to 65 indicates the depth of sediment samples (collected at 5-cm intervals) from 0–5 to 65–70 cm. Error bars indicate standard deviation.
