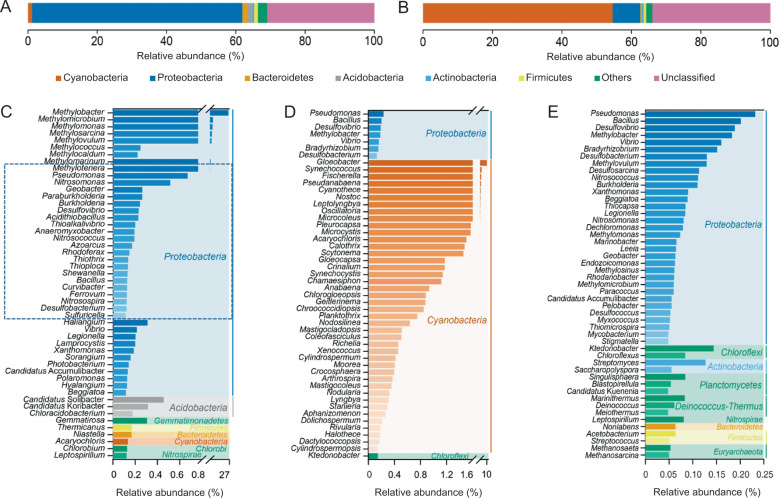
Fig. 6. Shotgun metagenomic sequencing data for total DNA from the sediment cores (the original sediment of 25–50 cm depth) and the heavy DNA from its 13CH4-incubated samples.
The phylum-level microbial community composition in the heavy DNA (A) and the original sediment (B), relative abundance of the top 50 genera in the heavy DNA (C), relative abundance of the top 50 genera in the original sediment (D), and the top 100 genera belonging to non-Cyanobacteria phyla in the original sediment (E). The genera affiliated with Proteobacteria in C identified with dotted rectangles may be associated with the metabolism of nitrogen, sulfide, metal (mainly iron) and electron transport based on reports in the literature (Supplementary Table S2). Taxonomic assignment of predicted genes was carried out using MEGAN (version 5.3) and subjected to BLASTX analysis using the NCBI-nr database (2016–09). The relative abundance of members of each taxonomic level was approximated by calculating the number of the gene reads affiliated with that taxonomic level to the total number of assembled reads per metagenome.