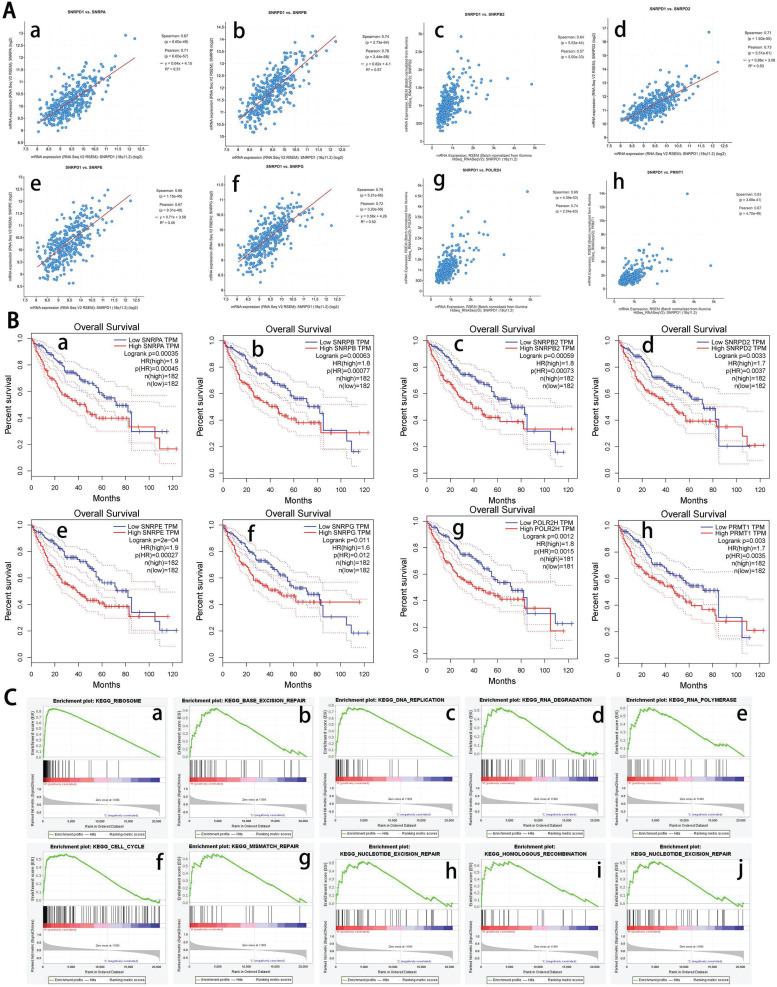
Figure 4.
Survival analysis of co-expressed genes interacted with SNRPD1and KEGG pathways analysis using GSEA. (A) Correlation between SNRPD1 and SNRPA (a, r=0.71), SNRPB (b, r=0.76), SNRPB2 (c, r=0.64), SNRPD2 (d, r=0.73), SNRPE (e, r=0.67), SNRPG (f, r=0.75), POLR2H (g, r=0.74), and PRMT1 (h, r=0.67), all P<0.05. (B) The survival analysis showed that the mRNA expression of SNRPA (a), SNRPB (b), SNRPB2 (c), SNRPD2 (d), SNRPE (e), SNRPG (f), POLR2H (g), and PRMT1 (h) were significantly related to the OS in HCC patients (all P<0.05). (C) The main enriched KEGG pathways of SNRPD1 using GSEA. RIBOSOME (a), BASE EXCISION REPAIR (b), DNA_REPLICATION (c), DNA REPLICATION (d), RNA POLYMERASE (e), CELL CYCLE (f), MISMATCH REPAIR (g), NUCLEOTIDE EXCISION REPAIR (h), HOMOLOGOUS RECOMBINATION (i), BLADDER CANCER (j).