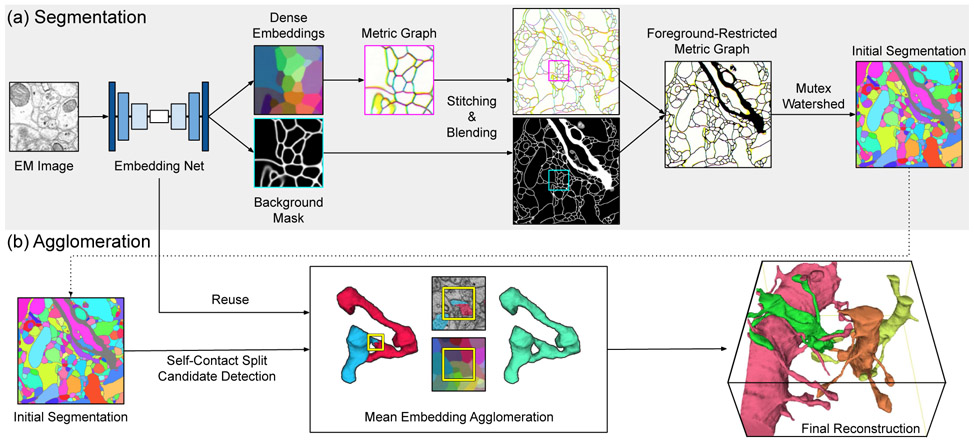
Fig. 1.
Overview of the proposed method. Dense voxel embeddings generated by a convolutional net (Sec. III-A) are employed at two subtasks for 3D neuron reconstruction: (a) neuron segmentation via metric graph [16] (Sec. III-B) and (b) agglomeration based on mean embeddings (Sec. III-C). All graphics shown here (images and 3D renderings) are drawn from real data. Although depicted in 2D for clarity, each 2D image represents a 3D volume. To visualize embeddings, we used PCA to project the 24-dimensional embedding space onto the three-dimensional RGB color space. For brevity, we only visualize nearest neighbor affinities on the metric graph by mapping x, y, and z-affinity to RGB, respectively.