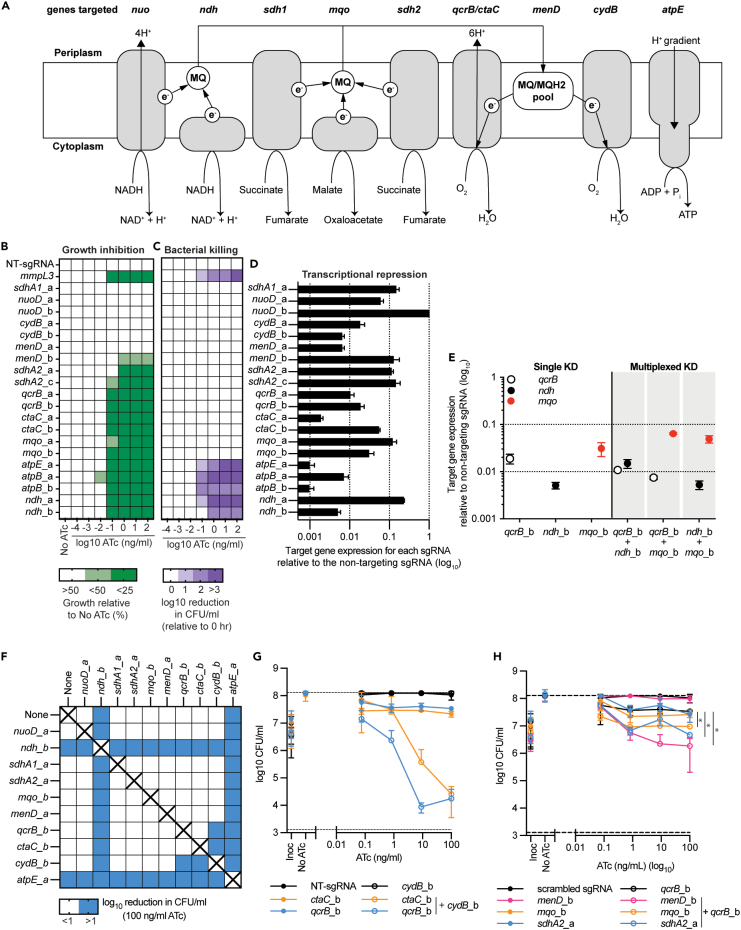
Figure 1.
Individual and simultaneous transcriptional repression of respiratory complexes in M. smegmatis
(A) Schematic of the genes targeted in this study and their functions in the mycobacterial electron transport chain. MQ: menaquinone, e−: electron, H+: proton. Arrowheads denote the direction of the catalyzed reaction, electron transfer or proton movement.
(B) Growth of M. smegmatis expressing sgRNAs from a starting OD600 of 0.005 in 96 well plates in a total volume of 100 μL with differing levels of ATc. Results are the mean ± standard deviation of four replicates, expressed relative to a no-ATc control. Increasing shades of green represent increasing levels of growth inhibition. White is representative of no growth impairment.
(C) Viability of M. smegmatis expressing sgRNAs as described for (B). Increasing shades of purple represent increasing levels of reduction in CFU/ml. White is representative of no reduction in CFU/ml. For B and C, results are mean ± standard deviation of four replicates with either non-targeting (NT) or mmpL3 targeting sgRNA that were used as negative and positive-bactericidal controls, respectively.
(D) mRNA levels of target genes following CRISPRi induction of stated sgRNA when induced with 100 ng/mL ATc and grown in LBT. Bar length represents the level of mRNA for each target gene with the stated sgRNA relative to a non-targeting sgRNA. 1 is representative of no change, whereas 0.1, 0.01, and 0.001 is representative of a 1, 2, and 3 log10 reduction in detectable mRNA, respectively. Results are the mean ± standard deviation of three technical replicates. (B–D) Labels a and b represent the guides a and b targeting each genetic target.
(E) Transcriptional analysis of qcrB, ndh, and mqo in either single or multiplexed knockdown strains. Results of targeted genes are expressed relative to a non-targeting control in the presence of 100 ng/mL ATc and are the mean ± SD of three technical replicates.
(F) Log10 reduction (0–26 h) in CFU/ml for single and multiplex strains in the presence of 100 ng/mL ATc. Blue shading represents a gene pair that when simultaneously repressed has at least a one-log reduction in CFU/ml. sgRNAs paired with “none” denote results from strains that express a single sgRNA. Combinations marked with “X” were not tested.
(G–H) CFU/ml plots of selected single and multiplexed KD against increasing concentrations of ATc. A strain expressing a non-targeting (NT) sgRNA is used as a negative control. (G) Expression of sgRNAs qcrB_b and ctaC alone or in combination with sgRNAs cydB_b. (H) Expression of sgRNAs menD_b, mqo_b, and sdhA2_a alone or in combination with sgRNA qcrB_b. For G and H, results are the mean ± SD of at least four biological replicates. ∗ denotes statistically significant difference with a p value of <0.05 between the average CFU/ml at 100 ng/mL for the multiplex sgRNA compared to the qcrB_b sgRNA as determined by an unpaired t test.