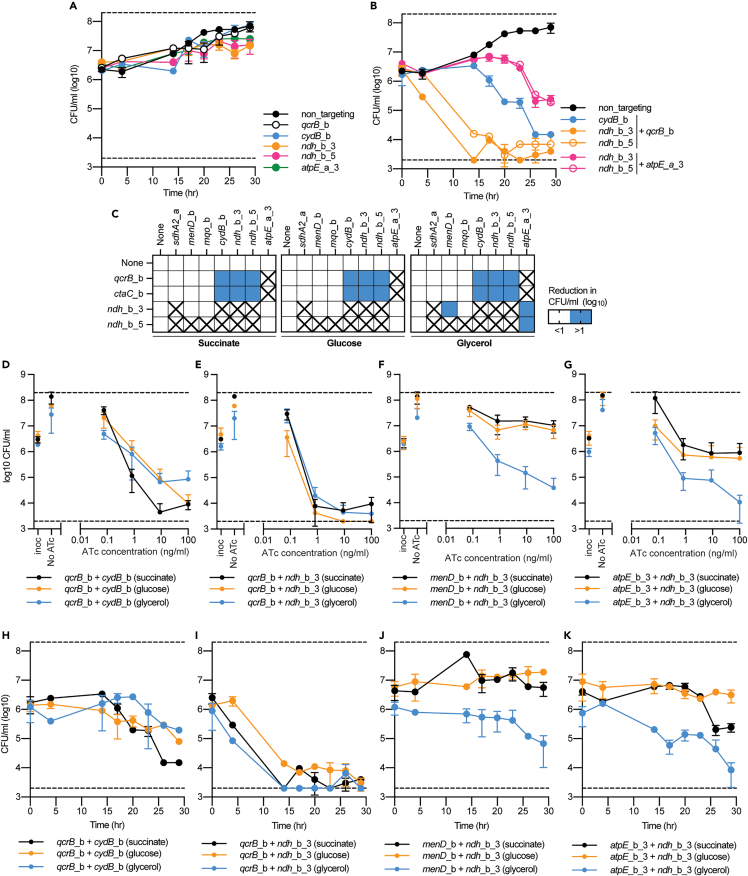
Figure 3.
Rate and magnitude of killing by synthetic lethal interactions is influenced by gene pair and growth on alternative carbon sources
(A–B) Time kill assays of either (A) single or (B) multiplexed gene pairs when grown in the presence of 100 ng/mL ATc with succinate as a sole carbon source. CFUs were taken at stated time points. For A-B, data is the mean ± SD of biological duplicates from a representative experiment.
(C) Log10 reduction (0–26 h) in CFU/ml for single and multiplex strains in the presence of 100 ng/mL ATc. Blue shading represents a gene pair that when simultaneously repressed has at least a 1 log greater reduction in CFU/ml. sgRNAs paired with “none” denote results from strains that express a single sgRNA. Boxes marked with an “X”, denoted gene pairs that were not analyzed on alternative carbon sources because they failed to show a synergistic or synthetic lethal interaction when previously tested (see Figures 1 and 2).
(D–G) CFU/ml plots of selected single and multiplexed KD against increasing concentrations of ATc. A strain expressing a non-targeting (NT) sgRNA is used as a negative control. Black, orange, and blue lines represent results when grown in succinate, glucose or glycerol, respectively. (D-G) Reduction in CFU/ml for M. smegmatis strains expressing the sgRNA (D-E) qcrB_b in combination with sgRNAs (D) cydB_b or (E) ndh_b_3, or (F-G) expression of the sgRNA ndh_b_3 in combination with (F) menD_b or (G) atpE_a_3. For D-G, results are the mean ± SD of at least four biological replicates.
(H–K) Kill curves for M. smegmatis strains expressing the sgRNA (H-I) qcrB_b in combination with sgRNAs (D) cydB_b or (E) ndh_b_3, or (F and G) expression of the sgRNA ndh_b_3 in combination with (F) menD_b or (G) atpE_a_3. For H-K, results are the mean ± SD two biological replicates from a representative experiment. Black, orange, and blue lines represent results when grown in succinate, glucose, or glycerol, respectively. For H-K, the succinate kill curve is the same as presented in panel 3B.