Figure 2.
Ex vivo and induced Tfh display distinct phenotypic landscapes and differentiation trajectories
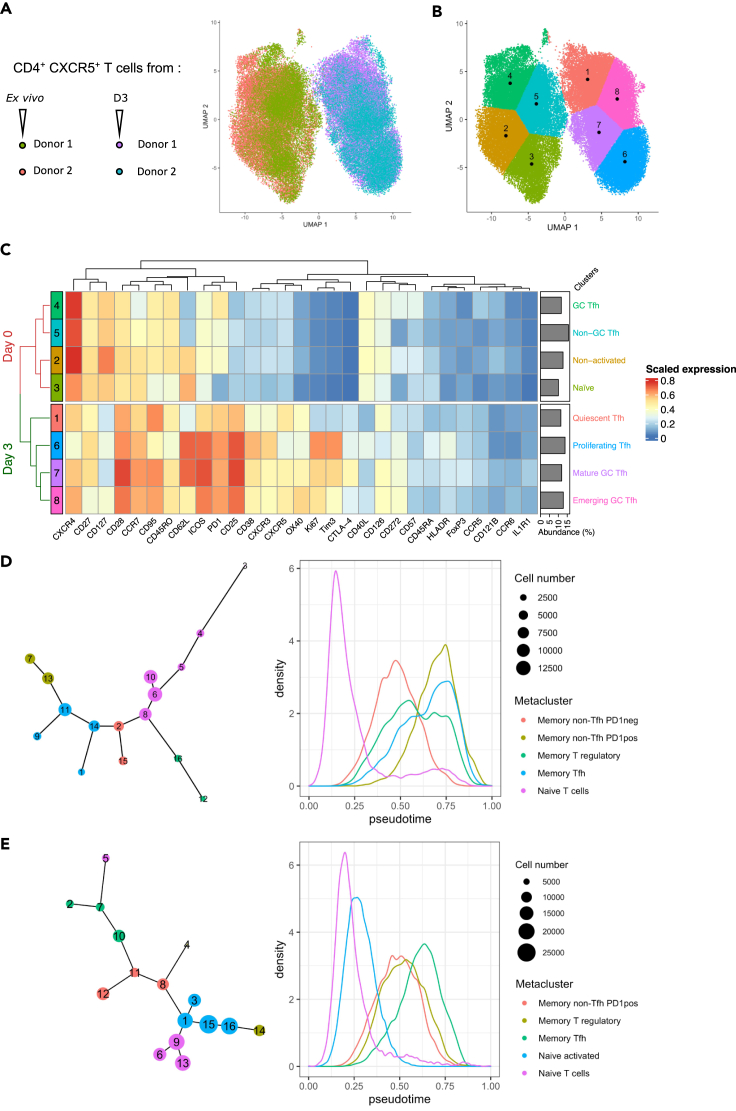
(A) Deep immunophenotyping was performed after 3 days of culture (n = 2 independent donors). CD4+ CXCR5+ T cell selection was based on the expression of CD45, CD8, CD11c, CD56, and CD19. The Uniform Manifold Approximation and Projection (UMAP) algorithm was used to represent the whole set of CD4+ CXCR5+ T cells in a multiparametric manner at D0 and D3 after antigenic stimulation of splenocytes from two donors.
(B) Projection of 8 clusters determined by k-means on the UMAP representation of D0 and D3 CD4+ CXCR5+ T cells.
(C) Heatmap representing the mean expression of 29 markers by 8 cell clusters and their relative abundance, defined among D0 and D3 CD4+ CXCR5+ T cells.
(D and E) Trajectory and pseudotime analysis on total D0 (D) and D3 (E) CD4+ T cells. Tree plots (left) show cluster trajectories, cell number, and metacluster assignment, and density plots (right) show the density of pseudotime across metaclusters.