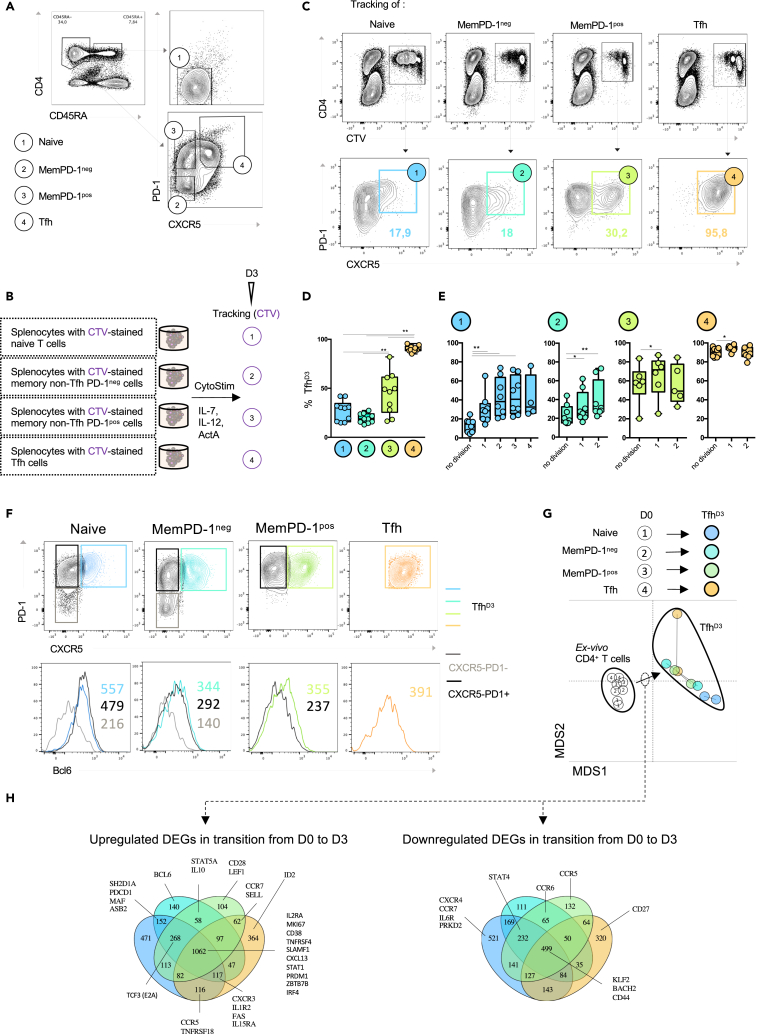
Figure 3.
Naive and memory CD4+ can orient toward a Tfh cell profile
(A) Four CD4+ T cell subsets are defined at day 0: (1) naive CD4+ CD45 RA+ T cells, (2) memory CD4+ CD45 RA- PD-1- T cells (MemPD-1neg), (3) memory CD4+ CD45 RA- PD-1+ T cells (MemPD-1pos), and (4) Tfh.
(B) At day 0, D4+ T cell subsets were sorted according to the gating strategy presented in (A). Isolated CD4+ T cell subsets were stained with cell trace violet (CTV) and mixed back into the negative splenocyte fraction. Stimulation and culture were next performed as previously described (Figure 1A).
(C) Representative flow plots of CTV tracking for each stimulated CD4+ T cell subset 3 days after antigenic stimulation (top) combined with flow cytometry analysis of CXCR5 and PD-1 expression among CTV+ cells at day 3 (bottom).
(D) Percentage of CXCR5+PD-1+ cells (TfhD3) among traced CD4+ T cell subsets after 3 days of antigenic stimulation.
(E) Percentage of TfhD3 cells according to the divisions of stimulated CD4+ T cell subsets.
(F) Representative histograms of Bcl6 expression for TfhD3 (colored line) compared with CXCR5−PD-1+ (black line) and CXCR5−PD-1- (gray line) deriving from respective CD4+ T cell subsets naive (blue), MemPD-1neg (turquoise blue), MemPD-1pos (green), and Tfh (orange).
(G) RNA sequencing was performed on CD4+ T cell subsets (Day 0) and the corresponding derived TfhD3 counterparts (n = 2). Multidimensional scaling was used to better visualize transcriptomic proximity of different CD4+ T cells.
(H) Venn diagrams were used to highlight Tfh-associated genes (Table 2) among differentially expressed genes that were shared during transition from D0 CD4+ T cell subsets to their TfhD3 counterparts (D and E). Each symbol represents an individual donor. A Wilcoxon matched pairs test was performed; ∗, p < 0.05; ∗∗, p < 0.005.