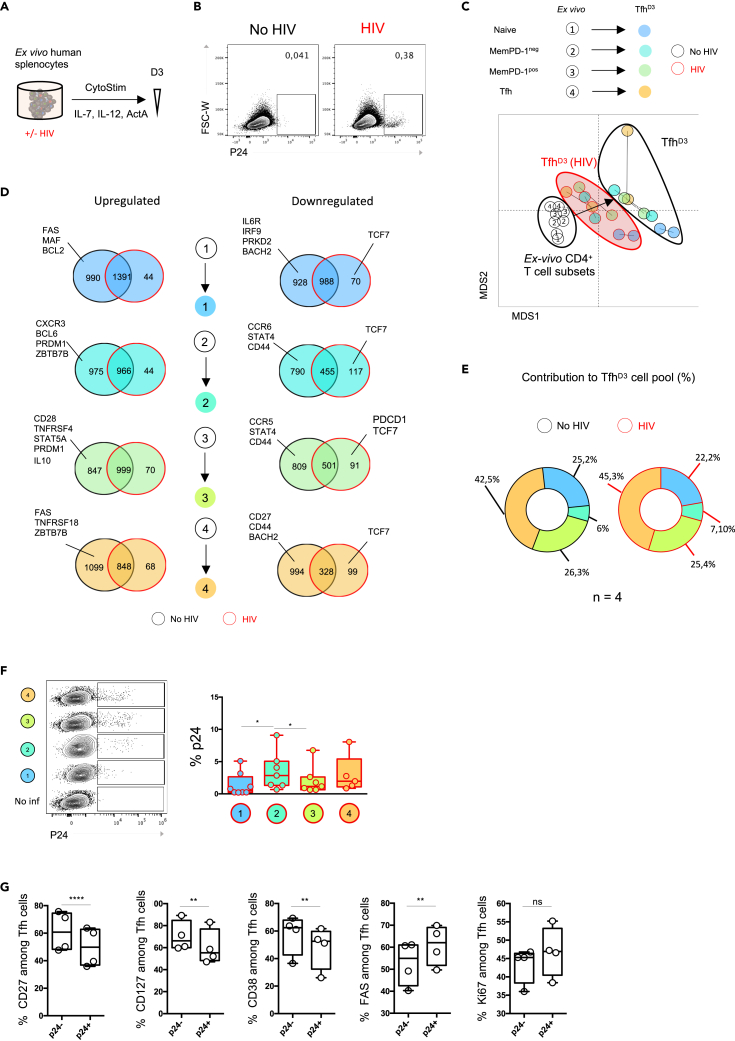
Figure 5.
HIV infection shapes Tfh cell differentiation and functions
(A) Splenocytes were stimulated according to the previously described protocol in the presence of HIV lab strain (Yu2b).
(B) Representative flow plots of p24 staining among splenocytes after 3 days of culture with HIV or not.
(C) RNA sequencing was performed on TfhD3 cells derived from distinct CD4+ T cell subsets with and without HIV. Multidimensional scaling was performed to visually cluster different CD4+ T cell populations based on their transcriptional profile (8,593 genes).
(D) RNA sequencing was performed on Tfh derived from each CD4+ T cell subset in the presence of HIV-1 infection or not. Differentially expressed genes were analyzed between Tfh and their original counterpart. Venn diagram representing (un)shared downregulated and upregulated genes. Genes specifically involved in Tfh cell biology were analyzed (referred to in Table 1).
(E) Contribution of each CD4 T cell subset to total Tfh generated after 3 days of splenocyte culture (%). Data are plotted as the mean percentage contribution of each ex vivo CD4+ T cell subset: (1) naive CD4+ T cells, (2) (3) memPD-1neg/pos, and (4) Tfh to total TfhD3 cells after splenocyte culture.
(F) Representative flow plot of p24 staining in Tfh derived from distinct CD4+ T cell subsets.
(G) Frequency of CD127-, CD27-, CD38-, FAS-, and ki67-positive cells among Tfh that are infected (p24pos) or not (p24neg). Each symbol (A–E) represents an individual donor. (E and G) A paired Student’s t test was performed, ∗p < 0.05, ∗∗p < 0.01. (F and G) A Wilcoxon matched pairs test was performed; ∗, p < 0.05; ∗∗, p < 0.005; ∗∗∗, p < 0.001.