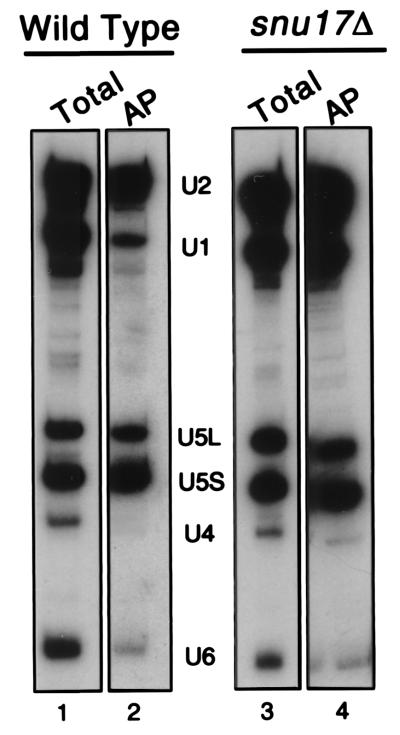
FIG. 7.
Deletion of SNU17 inhibits the dissociation of U1 and U4 from the spliceosome. Splicing-reaction mixtures were incubated with actin pre-mRNA that was transcribed in the presence of biotinylated UTP to incorporate an affinity label. The reaction mixtures were incubated for 20 min at 23°C. Then 5 μl was withdrawn and kept for analysis of total RNAs (Total; lanes 1 and 3). The remaining 120 μl was treated with heparin and then sedimented by centrifugation on a 10 to 30% glycerol gradient. rRNAs from yeast were run on a parallel gradient as sedimentation markers. Fifteen fractions (160 μl each) were collected, and four-fifths of each fraction was affinity purified with streptavidin-agarose (AP = affinity purification of a fraction of the 40S region of the gradient corresponding to fraction 15, the spliceosome [lanes 2 and 4]). After extensive washing, precipitated snRNAs were assayed by Northern blotting.