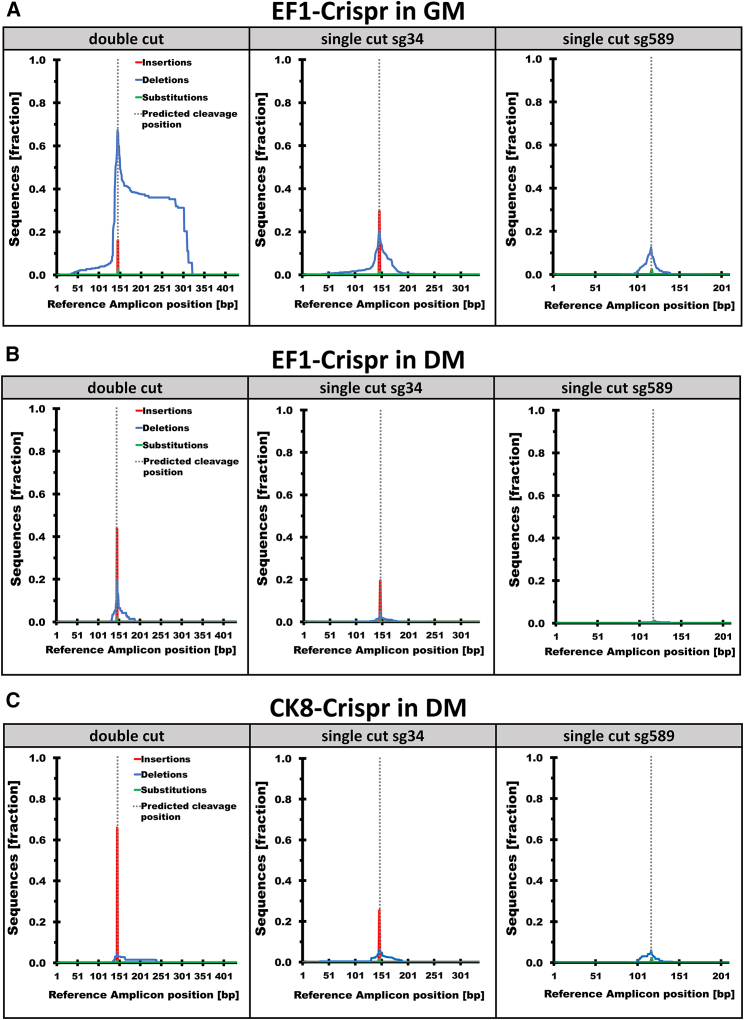
Figure 5.
Amplicon sequencing analysis reveals higher editing accuracy in differentiated cells than in proliferating cells
Genomic DNA was extracted from DM1 patient-derived proliferating cells in GM infected with (A) EF1-Crispr (n = 3) and differentiated cells in DM infected with (B) EF1-Crispr (n = 3) or (C) CK8-Crispr (n = 2). The precision of CRISPR/Cas9 activity was analyzed by amplicon sequencing of three possible editing events. For the double cut (simultaneous cut of both sgRNAs), primers located upstream of sg34 and downstream of sg589 were used. Single-cut events were examined using primers flanking the target site for sg34 or sg589 (single-cut sg34, single-cut sg589). The fraction of insertions, deletions, and substitutions resulting from this analysis was obtained by comparison with the reference sequence (Reference Amplicon, expected sequence derived from perfect rejoining at single- and double-cut sites).