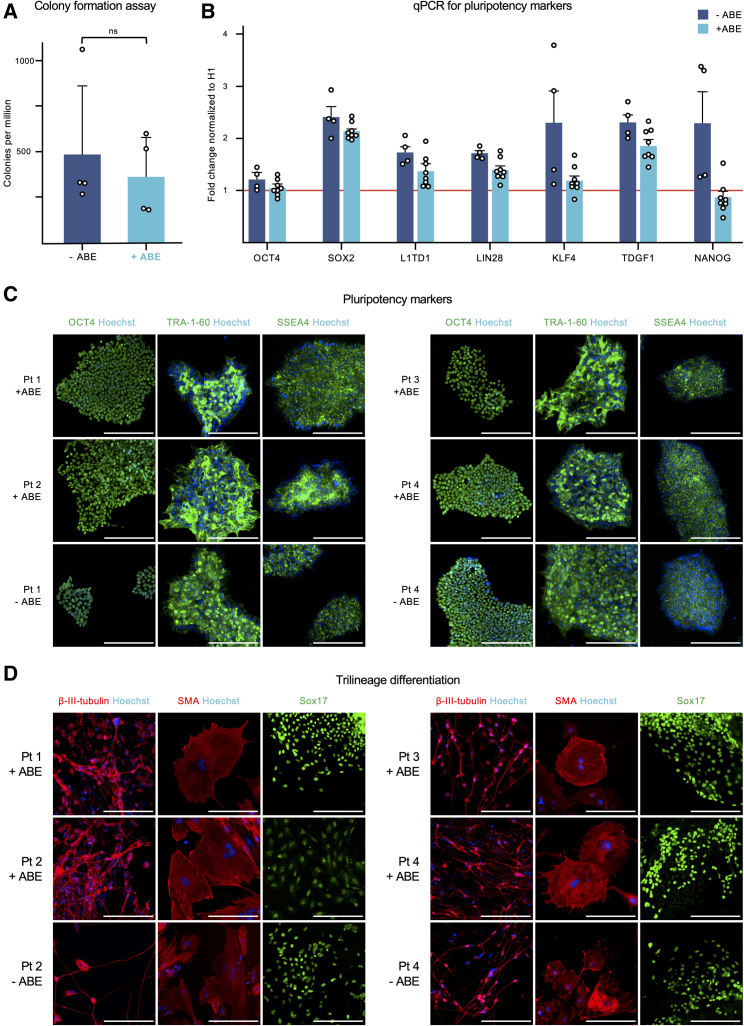
Figure 2.
Phenotypic analysis of monoclonal hiPSCs derived from simultaneously edited and reprogrammed primary fibroblasts
(A) Colony formation assay after the electroporation of 1 million fibroblasts with the three reprogramming vectors in the presence (+ABE, n = 4) or absence (−ABE, n = 4) of the ABEmax RNA construct and the sgRNA. Each point represents an independent assay from each of the four patient-derived fibroblast populations. Data are represented as mean + standard deviation.
(B) qPCR results for the expression levels of OCT4, SOX2, L1TD1, LIN28, KLF4, TDGF1, and NANOG in one non-ABE (−ABE, n = 4) and two base-edited (+ABE, n = 8) hiPSC lines per patient. The eight independently edited hiPSC lines were obtained through reprogramming plus simultaneous adenine base editing. The expression levels were normalized to those of the commercial H1 human embryonic stem cells, illustrated by the orange line. Each point represents an independent hiPSC line. Data are represented as mean + SEM. There were no significant differences between −ABE and +ABE.
(C) Immunofluorescence staining of four representative hiPSC lines base edited at the targeted adenine (+ABE) and their non-ABE controls (−ABE) for pluripotency markers OCT4, TRA-1-60, and SSAE4.
(D) Immunofluorescence staining of embryoid bodies derived from representative hiPSC lines base edited at the targeted adenine (+ABE) and their non-ABE controls (−ABE). β-III-Tubulin (ectoderm), smooth muscle actin (SMA) (mesoderm), and Sox17 (endoderm). NOTCH3: patient 1 and patient 2. LDLR: patient 3 and patient 4. Hoechst, in blue, is a nuclear marker. The white bar represents 200 μm.