Figure 5.
RNA-seq analysis to discover factors that might regulate the engraftability of skeletal myogenic progenitors
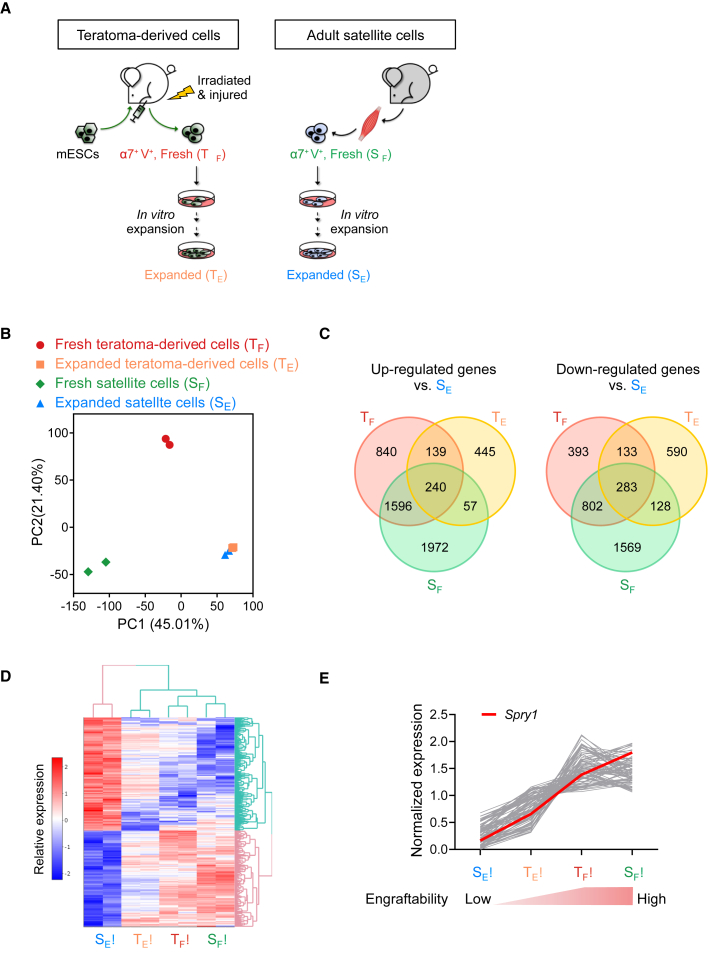
(A) Schematic of samples used for RNA-seq analysis: freshly isolated (TF) and expanded (TE) teratoma-derived skeletal myogenic progenitors and freshly isolated (SF) and expanded (SE) adult satellite cells (n = 2 biological replicates from each group).
(B) Principal-component analysis. Note that the transcriptomes of the three engraftable cell populations (TF, TE, and SF) are relatively spread out and the two expanded cell populations (TE and SE) are very close to each other.
(C) Venn diagrams showing differentially expressed genes (fold change >1.25, adjusted p < 0.05) that are commonly upregulated (left) or downregulated (right) in the three engraftable cell populations (TF, TE, and SF) versus the non-engraftable cells (SE).
(D) Heatmap showing the 240 upregulated and 283 downregulated genes from (C).
(E) Expression of upregulated genes with a Pearson correlation coefficient of >0.9 (gray lines) in the four cell populations. Expression of Spry1 is shown in red. See text for details.
See also Figure S5 and Tables 1 and S2.