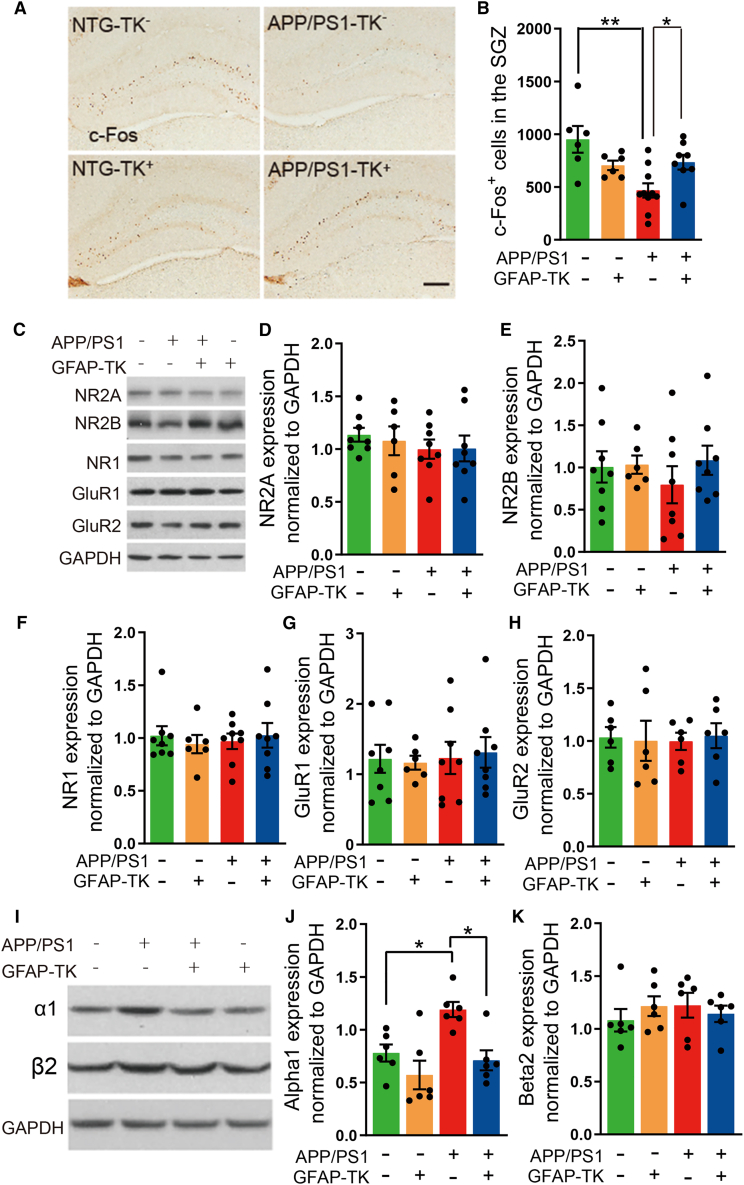
Figure 5.
Remodeling in the hippocampus after ablation of aNSCs
(A) Photomicrographs of c-Fos immunostaining in the DG of NTG-TK−, APP/PS1-TK−, APP/PS1-TK+, and NTG-TK+ mice after GCV treatment. Scale bar, 200 μm.
(B) Quantification of the numbers of c-Fos+ cells in the DG granule layer (NTG-TK−, n = 6; APP/PS1-TK−, n = 10; APP/PS1-TK+, n = 8; NTG-TK+, n = 6). Two-way ANOVA: genotype × treatment, F(1,26) = 10.07, p = 0.0038; genotype, F(1,26) = 0.0158, p = 0.9010; treatment, F(1,26) = 7.857, p = 0.0094; ∗p < 0.05, ∗∗p < 0.01 with Bonferroni post hoc test, data are represented as mean ± SEM.
(C) Protein bands of NMDARs (NR2A, NR2B, and NR1) and AMPARs (GluR1 and GluR2) in the hippocampus; GAPDH severed as the loading control.
(D–H) Quantification of the levels of NR2A, NR2B, NR1, GluR1, and GluR2 in the hippocampus of NTG-TK−, APP/PS1-TK−, APP/PS1-TK+, and NTG-TK+ mice after GCV treatment (n = 6 mice per group). Two-way ANOVA with Bonferroni post hoc test, data are represented as mean ± SEM.
(I) Protein bands of α1 and β2 GABAA receptor subunits in the hippocampus, GAPDH severed as the loading control.
(J) Quantification of the levels of α1 GABAA receptor subunit in the hippocampus of 4-month-old NTG-TK−, APP/PS1-TK−, APP/PS1-TK+, and NTG-TK+ mice after GCV treatment (n = 6 mice per group). Two-way ANOVA: genotype × treatment, F(1,20) = 1.904, p = 0.1829; genotype, F(1,20) = 12.21, p = 0.0023; treatment, F(1,20) = 7.796, p = 0.0113; ∗p < 0.05 with Bonferroni post hoc test, data are represented as mean ± SEM.
(K) Quantification of the levels of β2 GABAA receptor subunit in the hippocampus of 4-month-old NTG-TK−, APP/PS1-TK−, APP/PS1-TK+, and NTG-TK+ mice after GCV treatment (n = 6 mice per group). Two-way ANOVA: genotype × treatment, F(1,20) = 1.144, p = 0.2976; genotype, F(1,20) = 0.0674, p = 0.7979; treatment, F(1,20) = 0.1242, p = 0.7282; with Bonferroni post hoc test, data are represented as mean ± SEM.