Figure 6.
Enriched environment induced memory recovery and enhanced adult neurogenesis in APP/PS1 mice
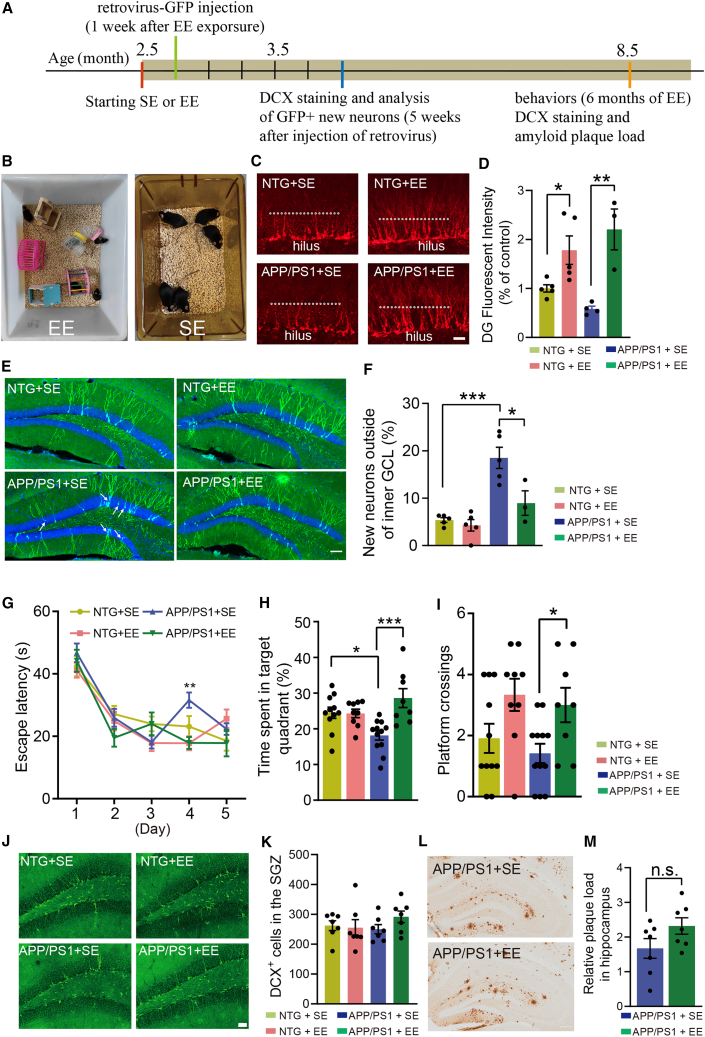
(A) Timeline for the experiments.
(B) Setup of the enriched environment (EE) and standard environment (SE).
(C) Representative photomicrographs of DCX+ cells in the DG of 4-month-old mice (6 weeks after exposure to EE). White dotted lines indicate the border between granule layer and molecular layer in the DG. Scale bar, 50 μm.
(D) Quantification of DCX expression in the DG of mice as in (C), n = 5 (NTG +SE or EE), 4 (APP/PS1 + SE), and 3 (APP/PS1 + EE). Two-way ANOVA: genotype × treatment, F(1,13) = 3.236, p = 0.0953; genotype, F(1,13) = 26.34, p = 0.0002; treatment, F(1,13) = 4.884e-005, p = 0.9945; ∗p < 0.05, ∗∗p < 0.01 with Bonferroni post hoc test, data are represented as mean ± SEM.
(E) Representative photomicrographs of newborn neurons labeled with retrovirus-GFP (5 weeks after the virus injection). White arrows indicate the newborn neurons located outside of the inner granule layer in the DG. Scale bar, 100 μm.
(F) Quantification of newborn neurons located in the middle or outer granule layer, n = 5 (NTG + SE or EE), 5 (APP/PS1 + SE), and 3 (APP/PS1 + EE). Two-way ANOVA: genotype × treatment, F(1,14) = 6.085, p = 0.0272; genotype, F(1,14) = 9.702, p = 0.0076; treatment, F(1,14) = 27.16, p = 0.0001; ∗p < 0.05, ∗∗∗p < 0.001 with Bonferroni post hoc test, data are represented as mean ± SEM.
(G) Learning curve in the Morris water maze test, n = 11 (NTG +SE), 9 (NTG + EE), 12 (APP/PS1 + SE), and 8 (APP/PS1 + EE). Day 4: two-way ANOVA, genotype × treatment, F(1,36) = 1.760, p = 0.1930; genotype, F(1,36) = 10.53, p = 0.0025; treatment, F(1,36) = 1.826, p = 0.1851; ∗∗p < 0.01 with Bonferroni post hoc test, data are represented as mean ± SEM.
(H) Time spent in the target quadrant during the probe trial of the Morris water maze test, n = 11 (NTG +SE), 9 (NTG + EE), 12 (APP/PS1 + SE), and 8 (APP/PS1 + EE). Two-way ANOVA: genotype × treatment, F(1,36) = 10.24, p = 0.0029; genotype, F(1,36) = 9.574, p = 0.0038; treatment, F(1,36) = 0.3785, p = 0.5423; ∗p < 0.05, ∗∗∗p < 0.001 with Bonferroni post hoc test, data are represented as mean ± SEM.
(I) The number of platform crossings during the probe trial of the Morris water maze test, n = 11 (NTG +SE), 9 (NTG + EE), 12 (APP/PS1 + SE), and 8 (APP/PS1 + EE). Two-way ANOVA: genotype × treatment, F(1,36) = 0.0293, p = 0.8651; genotype, F(1,36) = 10.47, p = 0.0026; treatment, F(1,36) = 0.7889, p = 0.3803; ∗p < 0.05 with Bonferroni post hoc test, data are represented as mean ± SEM.
(J) Representative photomicrographs of DCX+ cells in the DG of 9-month-old mice (6.5 months after exposure to EE). Scale bar, 50 μm.
(K) Quantification of DCX+ cells in the DG of mice as in (K), n = 7. Two-way ANOVA: genotype × treatment, F(1,24) = 1.436, p = 0.2426; genotype, F(1,24) = 0.7201, p = 0.4045; treatment, F(1,24) = 0.3715, p = 0.5479; with Bonferroni post hoc test, data are represented as mean ± SEM.
(L) Representative photomicrographs of amyloid deposition (staining with 3D6) in the hippocampus of 9-month-old APP/PS1 mice (6.5 months after exposure to EE or SE). Scale bar, 100 μm.
(M) Quantification of the amyloid plaque load in the hippocampus of APP/PS1 mice as shown in (L). Unpaired t test, data are represented as mean ± SEM. See also Figures S5–S7.