Figure 4.
Mass cytometry of hiPSCs and hESCs and comparison of manual and automated cell culture
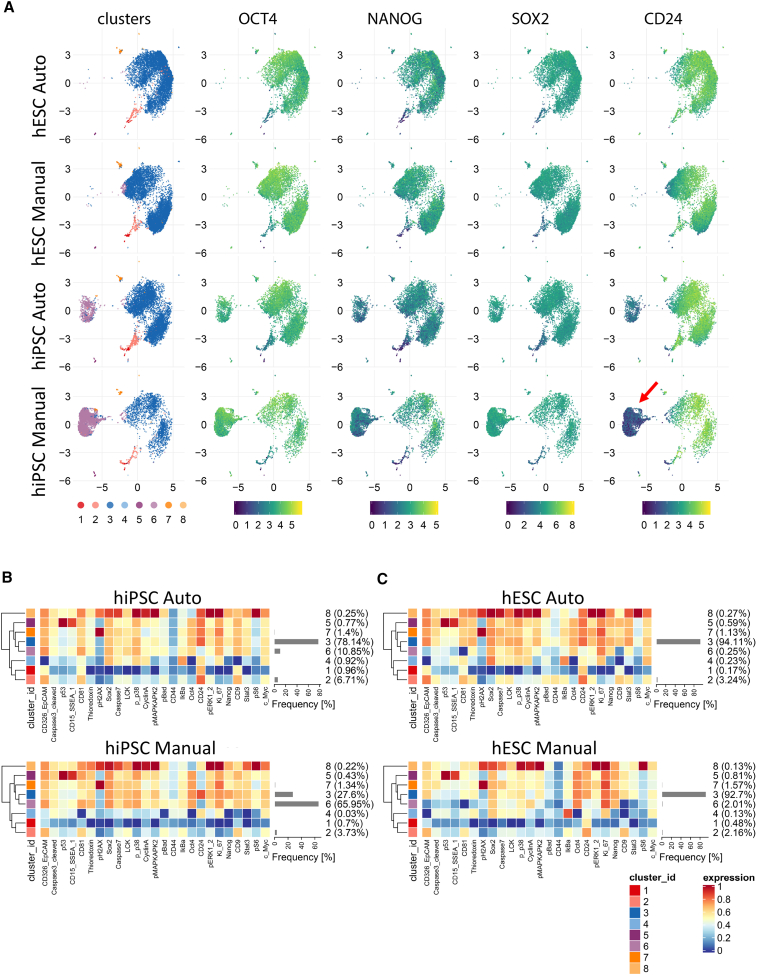
(A) Uniform manifold approximation and projection (UMAP) plots showing subpopulations of cells within each group organized into eight clusters identified by FlowSOM and ConsensusClusterPlus algorithms. Cluster 6 was prominent in hiPSCs (LiPSC-GR1.1) when cultured manually and its representation was mitigated by automated culture. Core pluripotency markers OCT4, NANOG, and SOX2 were expressed at similar levels across clusters. Surface-antigen CD24 was expressed at a considerably higher level in cluster 6 in hiPSCs cultured manually (red arrow).
(B) Heatmaps comparing protein expression levels for each analyzed marker in individual clusters and the abundance of the clusters within the hiPSC populations (LiPSC-GR1.1) cultured manually or by automation. Manual culture led to a large proportion of CD24-negative cells, 66% versus 11% in automated culture.
(C) Heatmaps of protein expression levels and cluster abundances in hESCs (WA09) after manual and automated cell culture. The abundance of the major cluster 3 was similar in both culture conditions, and CD24-negative cluster 6 was represented at a negligible level. UMAP plots were constructed from 8,000 single cells per sample (n = 4 independent experiments) obtained from two independent cell lines (A–C). CyTOF data were analyzed using a modified CyTOF workflow (Robinson et al., 2017).