Figure 5. Experimental approaches for phasing SNVs to establish haplotype.
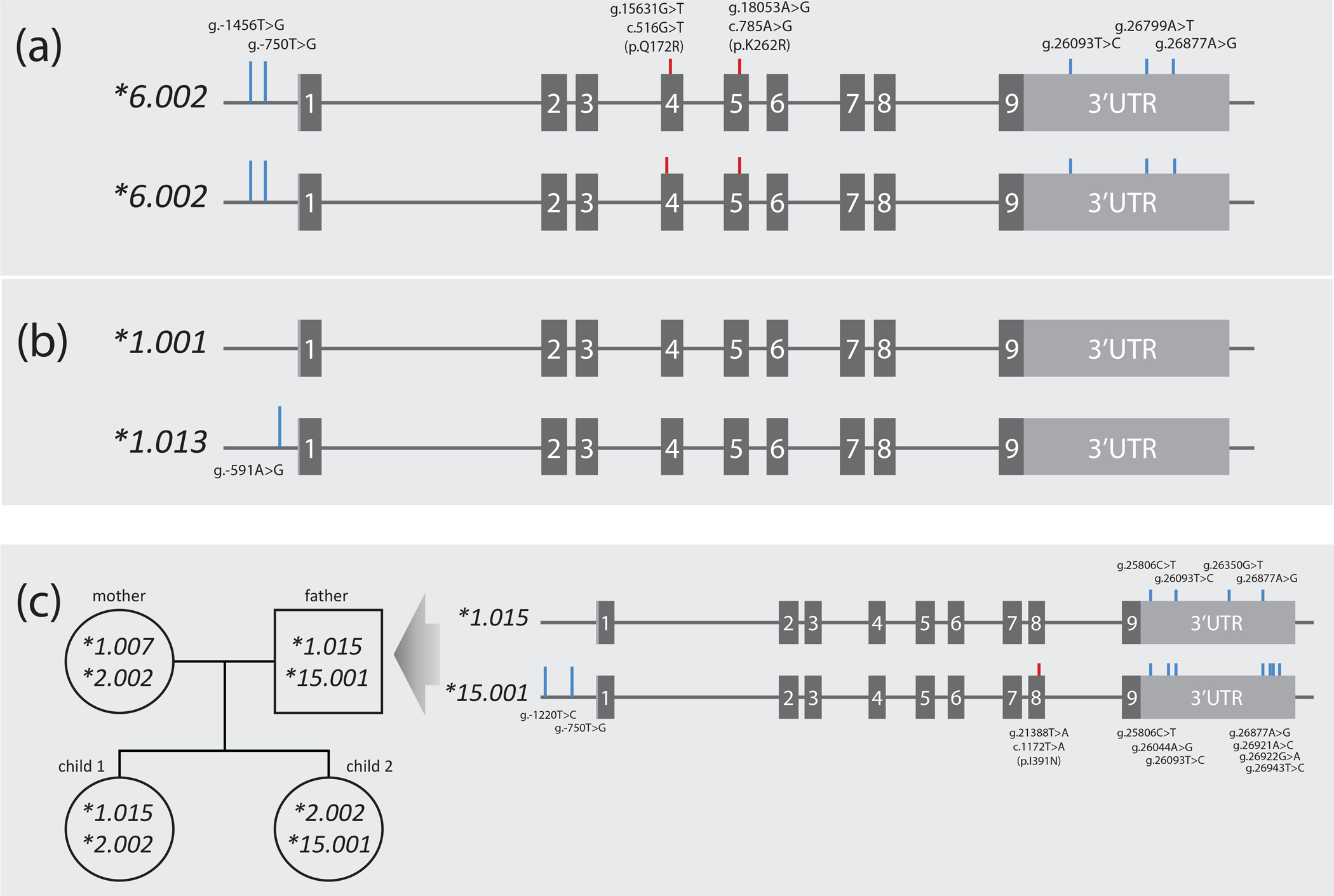
Panel A represents samples NA12003 and NA19176 both of which were homozygous for eight SNVs. Two are CYP2B6*6 core SNVs (c.516G>T (pQ172R) and c.785A>G (p.K262R); red lines) while the remaining SNVs sub-classify this haplotype as *6.002 (blue lines). Homozygosity across all SNVs unequivocally informs haplotype. Panel B depicts NA12156 which is heterozygous for a single SNV and thus unequivocally informs the haplotype of this novel suballele which was designated *1.013. Panel C exemplifies how haplotype can be inferred by inheritance (the example shown was part of a PharmVar submission). Haplotypes were determined using WGS data for a family with two offspring. Inferred haplotypes were corroborated with 10X Genomics Long-Read sequence technology for all family members. The CYP2B6*1.015 and *15.001 alleles of the father are detailed as indicated by the arrow. CYP2B6*1.007 found in the mother was found homozygous in NA18544, NA17227 and NA18952 (not shown) further supporting the use of these technologies to inform CYP2B6 haplotype.
