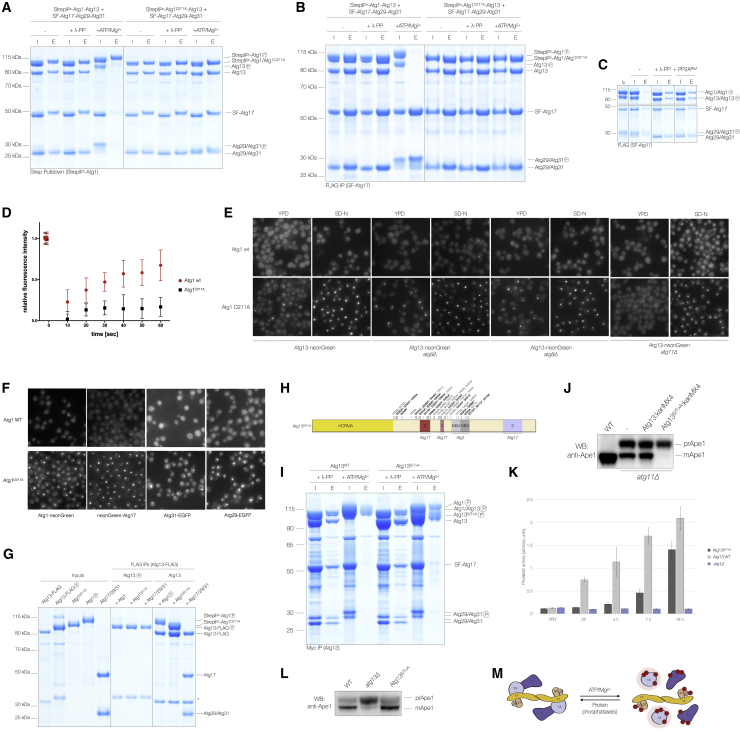
Figure 3.
Atg1-mediated phosphorylation of Atg13 dissociates the Atg1 complex
(A) StrepII2x-Atg1-Atg13 or catalytically inactive StrepII2x-Atg1D211A-Atg13 was mixed with Atg17-Atg29-Atg31 (input: I) and incubated with ATP/Mg2+ or λ-PP. Atg1 was immobilized using StrepTactin resin. Atg1 and co-purifying proteins were eluted (E) and analyzed by SDS-PAGE.
(B) Atg1-Atg13 or Atg1D211A-Atg13 was mixed with SF-Atg17-Atg29-Atg31 (input: I) and incubated with or without ATP/Mg2+ or λ-PP. Atg17 and co-purifying proteins were eluted (E) and analyzed by SDS-PAGE.
(C) The Atg1-Atg13-Atg17-Atg29-Atg31 complex was incubated with ATP/Mg2+ to trigger complex dissociation (I0). ATP was depleted using apyrase and samples were incubated with either λ-PP, PP2ARts1, or no PP (I). SF-Atg17 was immobilized and pull-down elutions (E) were analyzed by SDS-PAGE.
(D) Fluorescence recovery after photobleaching (FRAP) experiments monitoring Atg13-neonGreen in atg11Δ cells expressing either WT Atg1 or Atg1D211A. Quantification shows the relative fluorescence intensities after bleaching the Atg13-neonGreen signal as a function of time.
(E) Fluorescence microscopy analysis of WT, atg8Δ, atg9Δ, or atg11Δ cells expressing Atg13-neonGreen in the presence of WT Atg1 or Atg1D211A. Cells were exponentially grown in nutrient-rich YPD medium or nitrogen starved for 4 h. Scale bar, 5 μm.
(F) Fluorescence microscopy images of nitrogen-starved yeast expressing Atg1-neonGreen, Atg1D211A-neonGreen, or neonGreen-Atg17, Atg29-EGFP, or Atg31-EGFP in WT or atg1D211A cells. Scale bar, 5 μm.
(G) Atg13 was phosphorylated using substoichiometric amounts of Atg1. ATP was depleted using apyrase and phosphorylated, and non-phosphorylated Atg13 was immobilized. Autophosphorylated Atg1, Atg1D211A, or Atg17-Atg29-Atg31 were added, and Atg13 and co-purifying proteins were eluted and analyzed by SDS-PAGE. Asterisk, Atg13 truncation.
(H) Serines and threonines mutated to alanine in the Atg13S/T>A mutant are shown with respect to the N-terminal HORMA domain, Atg17 binding sites, and MIT-interacting motifs (MIMs). In vivo phosphorylation sites are marked by asterisks with putative Atg1-dependent phosphorylation sites underlined. Serines highlighted in black are phosphorylated by recombinant Atg1 in vitro.
(I) Atg13WT or Atg13S/T>A were mixed with Atg1 and Atg17-Atg29-Atg31 (I) and incubated with either ATP/Mg2+ or λ-PP. Atg13 and Atg13S/T>A were immobilized and elutions (E) were analyzed by SDS-PAGE.
(J) WT and atg11Δ cells expressing either Atg13WT or Atg13S/T>A were nitrogen starved (7 h), and processing of precursor Ape1 (prApe1) to its mature form (mApe1) was monitored by western blotting (WB).
(K) Pho8Δ60 assay measuring bulk autophagy in atg1Δ strains and yeast expressing either Atg13WT or Atg13S/T>A. Cells were either exponentially grown in YPD medium or nitrogen starved for 2, 4, 7, or 18 h. Alkaline phosphatase activity was measured (n = 3) and plotted as relative Pho8Δ60 activity with standard deviation.
(L) The indicated yeast strains were exponentially grown in YPD medium and Cvt pathway-dependent Ape1 processing was monitored by WB.
(M) Model summarizing phosphorylation-dependent dissociation of the Atg1 complex.