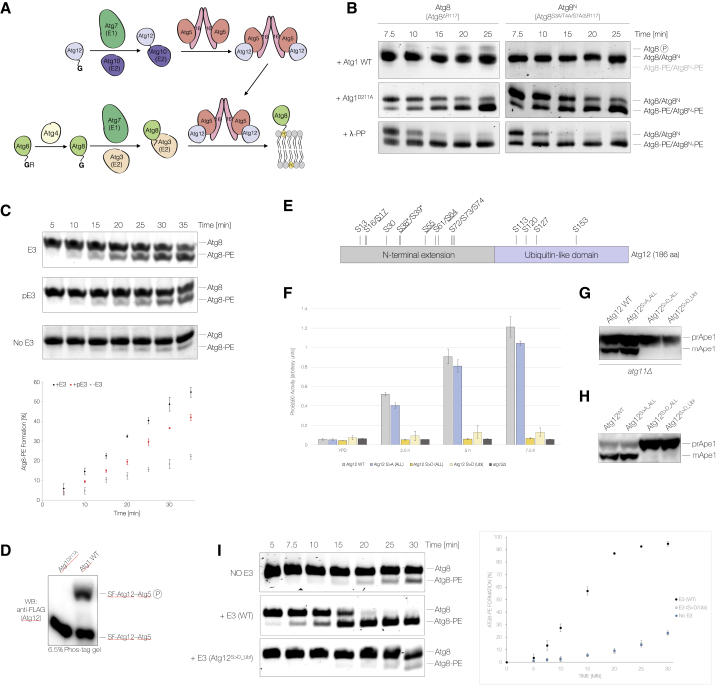
Figure 4.
Atg8 lipidation is inhibited by Atg1-mediated phosphorylation of the E3
(A) Schematic overview of Atg8 processing and lipidation.
(B) Recombinant Atg3, Atg7, and Atg12–Atg5-Atg16 were incubated with WT Atg1, Atg1D211A, or λ-PP in the presence of ATP/Mg2+ and PP inhibitors. Liposomes were added and lipidation reactions were started by the addition of Atg8 (Atg8ΔR117) or Atg8N (Atg8S3A/T4A/S7A/ΔR117). Samples were taken at indicated time points and analyzed by urea-SDS-PAGE and Sypro Ruby staining.
(C) Atg8 lipidation was monitored in the presence or absence of E3 and in the presence of an E3 pre-phosphorylated by Atg1 (pE3). Lipidation reactions were set up as illustrated in Figure S5G. Samples were taken and analyzed as in (B). The average lipidation ± standard deviation is plotted for each time point (n = 3; bottom panel).
(D) Endogenous SF-tagged Atg12 was purified from nitrogen-starved yeast expressing WT Atg1 or Atg1D211A. The electrophoretic mobility of the Atg12–Atg5 conjugate was compared by Phos-tag SDS-PAGE and WB.
(E) Schematic overview of S. cerevisiae Atg12 highlighting the N-terminal extension, Ubl domain, and Atg1-dependent in vitro phosphorylation sites. Asterisks indicate phosphorylation sites also identified in vivo (Hu et al., 2019; Lanz et al., 2021). Underlined sites were previously reported to be phosphorylated by Atg1 in vitro (Hu et al., 2019).
(F) Bulk autophagy was quantified in Atg12-, Atg12S>A_ALL-, Atg12S>D_ALL-, and Atg12S>D_Ubl-expressing or atg12Δ cells using the Pho8Δ60 assay. Cells were exponentially grown in YPD medium or nitrogen starved for 2.5, 5, or 7.5 h.
(G) Bulk autophagy was monitored in nitrogen-starved atg11Δ cells expressing WT Atg12, Atg12S>A_ALL, Atg12S>D_ALL, or Atg12S>D_Ubl. Ape1 processing was monitored by WB.
(H) The Cvt pathway was analyzed in cells expressing Atg12WT, Atg12S>A_ALL, Atg12S>D_ALL, or Atg12S>D_Ubl by monitoring Ape1 processing.
(I) Atg8 lipidation was monitored in the presence of an E3 containing either WT Atg12 or Atg12S>D_Ubl. Atg8 lipidation was analyzed as in (B).