Figure 5.
Atg8 lipidation is inhibited by Atg1-mediated phosphorylation of Atg3
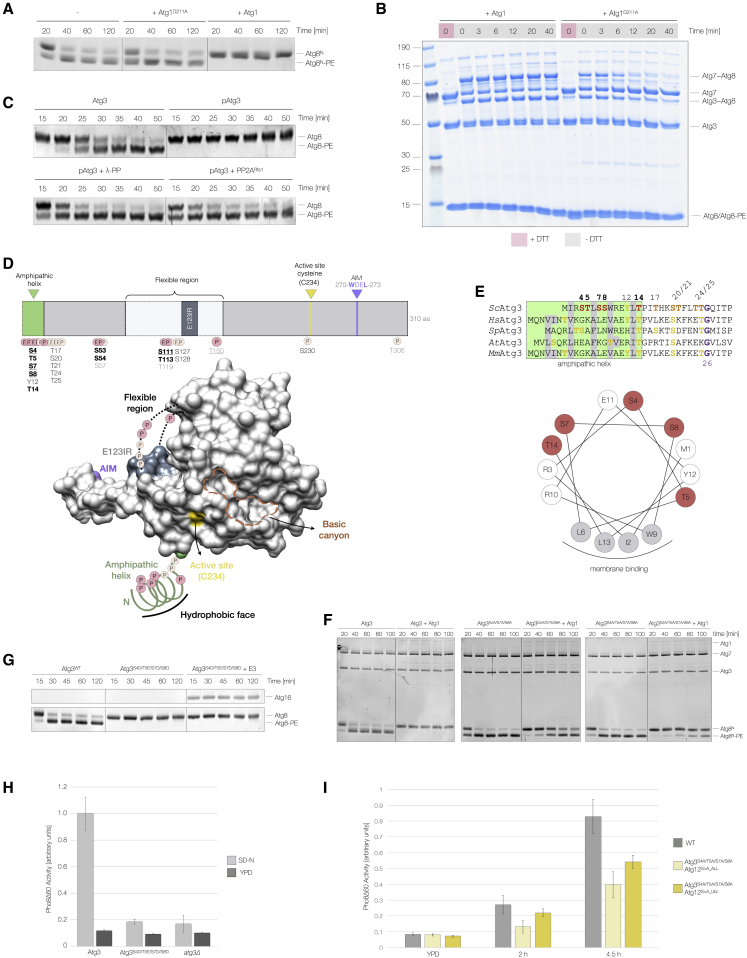
(A) Atg3 and Atg7 were incubated with either WT Atg1 or Atg1D211A in the presence of ATP/Mg2+. Liposomes were added and lipidation reactions were started by the addition of Atg8N. Lipidation was analyzed by urea-SDS-PAGE and Coomassie staining.
(B) Atg3, Atg7, and Atg8 were individually incubated with WT Atg1 or Atg1D211A and ATP/Mg2+ before combining to promote Atg3 charging. Liposomes were added to monitor Atg8 discharge in a time-dependent manner. Samples were analyzed by SDS-PAGE under reducing or non-reducing conditions.
(C) Atg8 lipidation assays were carried out using WT Atg3, Atg1-phosphorylated Atg3 (pAtg3), or λ-PP or PP2ARts1-treated pAtg3. Samples were taken at the indicated time points, and Atg8 lipidation was analyzed by urea-SDS-PAGE and Sypro Ruby staining.
(D) Schematic domain overview and surface representation of S. cerevisiae Atg3 (PDB: 2DYT) highlighting phosphorylation sites and unique functional elements, including the N-terminal amphipathic helix (green), the E1, E2, and E3 interacting region (E123IR), the active site cysteine, and the AIM. Sites phosphorylated in vivo and by Atg1 in vitro are highlighted in bold black. In vivo phosphorylation sites regulated by Atg1 are underlined. Phosphorylation sites detected either in vitro or in vivo are shown in gray and black, respectively.
(E) Sequence alignment of the Atg3 N terminus with Atg1-dependent in vitro and in vivo phosphorylation sites colored in red and orange. Residues in yellow indicate potential phosphorylation sites in other organisms. The helical wheel representation of the N-terminal amphipathic helix is shown below.
(F) Atg8N lipidation was compared in the presence or absence of Atg1 using either WT Atg3, Atg3S4A/S7A/S8A, or Atg3S4A/T5A/S7A/S8A. Atg7, Atg3, and Atg8N were separately incubated with or without Atg1 before starting the lipidation reaction. Samples were analyzed by urea-SDS-PAGE and Sypro Ruby staining.
(G) Atg8 lipidation was studied in the presence of Atg3 or Atg3S4D/T5E/S7D/S8D. In a separate reaction, the E3 was added to Atg3S4D/T5E/S7D/S8D-containing reactions. Atg8 lipidation was analyzed as in (F).
(H) Bulk autophagy was quantified in nitrogen-starved (4 h) WT Atg3, atg3Δ, and Atg3S4D/T5E/S7D/S8D-expressing cells using the Pho8Δ60 assay (n = 3).
(I) The Pho8Δ60 assay was used to quantify bulk autophagy in yeast co-expressing Atg3S4A/T5A/S7A/S8A with either Atg12S>A_ALL or Atg12S>A_Ubl (n = 3).