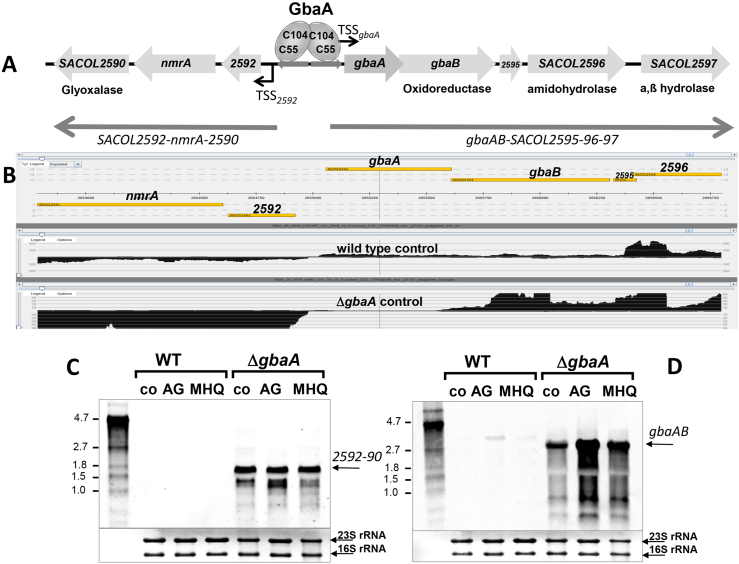
Fig. 2.
Deletion of gbaA results in derepression of transcription of the downstream gbaAB-SACOL2595-97 and upstream SACOL2592-90 operons. (A, B) Transcriptional organization of the divergent gbaAB-SACOL2595-97 and SACOL2592-90 operons in S. aureus. The upstream SACOL2592-nmrA-2590 operon encodes for a putative glyoxalase and NAD+-dependent epimerase/dehydratase (NmrA). The downstream gbaAB operon encodes for the GbaA repressor, a putative short chain oxidoreductase, an amidohydrolase and an a,ß hydrolase. (B) Both operons are negatively regulated by GbaA as displayed by the RNA-seq data of S. aureus COL WT and the gbaA mutant under control conditions using Read-Explorer. (C, D) Transcription of the SACOL2592-90 (C) and gbaAB-SACOL2595-97 operons (D) was analyzed in S. aureus COL WT and gbaA mutant strains before (co) and 30 min after treatment with 5 μg/ml AGXX® (AG) and 50 μM MHQ using Northern blots. Both operons remained inducible by AGXX® and MHQ stress in the gbaA mutant. The arrows point toward the transcript sizes of the gbaAB and SACOL2592-90 operons. The methylene blue bands denote the 16S and 23S rRNAs as RNA loading controls below the Northern blots. Band intensities of the gbaAB and SACOL2592-90 operon transcripts were quantified using ImageJ and the data are shown in Figs. S5A and B. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)